

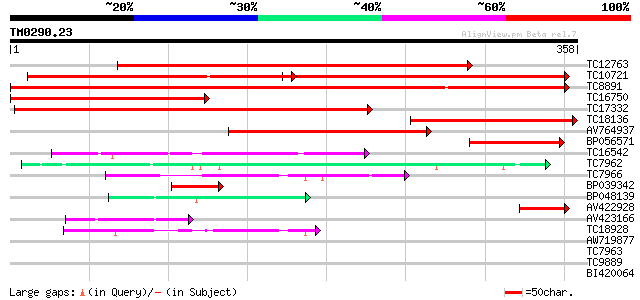
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.23
(358 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12763 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 449 e-127
TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehyd... 197 3e-94
TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Ara... 306 5e-84
TC16750 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 271 9e-74
TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehyd... 236 3e-63
TC18136 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 196 5e-51
AV764937 119 6e-28
BP056571 69 1e-12
TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, pa... 64 3e-11
TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 64 5e-11
TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, par... 56 8e-09
BP039342 49 1e-06
BP048139 48 2e-06
AV422928 47 6e-06
AV423166 46 8e-06
TC18928 similar to UP|Q43677 (Q43677) Auxin-induced protein, par... 45 2e-05
AW719877 37 0.004
TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 37 0.004
TC9889 similar to UP|Q9AYU1 (Q9AYU1) Quinone-oxidoreductase QR1 ... 35 0.019
BI420064 33 0.074
>TC12763 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (62%)
Length = 809
Score = 449 bits (1154), Expect = e-127
Identities = 223/224 (99%), Positives = 224/224 (99%)
Frame = +2
Query: 69 HEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDG 128
HEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDG
Sbjct: 68 HEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDG 247
Query: 129 KPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILG 188
KPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILG
Sbjct: 248 KPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILG 427
Query: 189 LGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYI 248
LGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYI
Sbjct: 428 LGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYI 607
Query: 249 IDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGR 292
IDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLG+
Sbjct: 608 IDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGK 739
>TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , complete
Length = 1297
Score = 197 bits (501), Expect(2) = 3e-94
Identities = 82/170 (48%), Positives = 122/170 (71%)
Frame = +3
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAARD SG+LSP+ F+ R G DDV IK+ +CGVCH+D+H +KND G + YP+VPGHE+
Sbjct: 117 GWAARDTSGVLSPFHFSRRENGDDDVAIKILFCGVCHSDLHTIKNDWGFTTYPVVPGHEI 296
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
VG V + G++VT+F VG+ VG G++V CK C C D+E YC + ++ YN Y +G T
Sbjct: 297 VGVVTKTGNNVTKFKVGDKVGVGVIVDSCKSCENCDQDLENYCPQPVYTYNSPY-NGTRT 473
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESG 181
GG+++ ++V Q++V++ P+ + + APLLCA +TVY P+ ++G+ E G
Sbjct: 474 NGGYSDFVVVHQRYVLQFPDNLPLDAGAPLLCAGITVYXPMKYYGMTEPG 623
Score = 164 bits (416), Expect(2) = 3e-94
Identities = 86/181 (47%), Positives = 118/181 (64%)
Frame = +1
Query: 173 SHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVS 232
S G + G G+ GLGG+GH+ + KA G VTVIS+S K+ EAIE LGAD++LVS
Sbjct: 598 SIMG*QSQGKHLGVAGLGGLGHVAIKFGKAFGLKVTVISTSPNKEAEAIEKLGADSFLVS 777
Query: 233 SDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGR 292
+D M+ A ++ YIIDT+ H L P L LLKL+GKL+ +G+ PL+ ++ GR
Sbjct: 778 TDPAKMKAALGTIHYIIDTISASHSLIPLLGLLKLNGKLVTVGLPGKPLELPVFPLVAGR 957
Query: 293 RSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKG 352
+ + GS G IKET+EMLEF + + + IE++ +D IN A ERL K DV+YRFV+DV
Sbjct: 958 KLVGGSHFGGIKETQEMLEFSAKHNIAADIELIKIDEINTAMERLSKADVKYRFVIDVAX 1137
Query: 353 S 353
S
Sbjct: 1138S 1140
>TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , complete
Length = 1636
Score = 306 bits (783), Expect = 5e-84
Identities = 154/354 (43%), Positives = 224/354 (62%), Gaps = 1/354 (0%)
Frame = +1
Query: 1 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 60
M S A +GWAA D SG+LSPY F+ R G DDVY+K+ +CGVC+ DV +N LG
Sbjct: 94 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 273
Query: 61 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKK-KIW 119
S YP VPGHE+ G V +VGS+V F+VG+ VG G V C+ C C +E +C K ++
Sbjct: 274 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 453
Query: 120 NYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKE 179
+N V DG T+GG+++ I+V +++ IP+ APLLCA +TVYSP+ + +
Sbjct: 454 TFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQ 633
Query: 180 SGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQ 239
G G++GLGG+GHM V KA G +VTV S+S KK+EA+ LGAD ++VSSD M+
Sbjct: 634 PGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMR 813
Query: 240 EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSF 299
A SLD+I+DT H +PY++LLK G L L+G ++ ++++G R+++GS
Sbjct: 814 ALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSG 990
Query: 300 IGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
+G KE +M+EF + IE+V + Y N+A +R+ K DV+YRFV+D++ S
Sbjct: 991 VGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 1152
>TC16750 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (34%)
Length = 442
Score = 271 bits (694), Expect = 9e-74
Identities = 126/126 (100%), Positives = 126/126 (100%)
Frame = +1
Query: 1 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 60
MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM
Sbjct: 64 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 243
Query: 61 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN 120
SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN
Sbjct: 244 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN 423
Query: 121 YNDVYV 126
YNDVYV
Sbjct: 424 YNDVYV 441
>TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 736
Score = 236 bits (603), Expect = 3e-63
Identities = 110/226 (48%), Positives = 151/226 (66%)
Frame = +1
Query: 4 LEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNY 63
+E + GWAARD SG+LSP+ F+ R TG DV KV YCG+CH+D+H +KN+ G S Y
Sbjct: 58 MEHPKKAFGWAARDSSGVLSPFKFSRRETGEKDVTFKVLYCGICHSDLHMIKNEWGSSVY 237
Query: 64 PMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYND 123
P+VPGHE+ G V EVGS V +F VG+ VG G +V C+ C +C ++E YC K I Y
Sbjct: 238 PLVPGHEIAGVVTEVGSKVQKFKVGDRVGVGCMVNSCRSCQSCADNLENYCPKMILTYAA 417
Query: 124 VYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLR 183
VDG T GG+++ ++ ++ F ++I + + E AP LCA +TVYSPL ++GL + GL
Sbjct: 418 KNVDGTITYGGYSDLMVSDEDFGIRISDNLPLEAAAPFLCAGITVYSPLKYYGLDKPGLH 597
Query: 184 GGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAY 229
G+ GLGG+GHM V A A+G VT IS+S KK EAIE + AD++
Sbjct: 598 VGVSGLGGLGHMAVKFAIALGAKVTAISTSPNKKTEAIEKV*ADSF 735
>TC18136 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (28%)
Length = 601
Score = 196 bits (498), Expect = 5e-51
Identities = 99/105 (94%), Positives = 102/105 (96%)
Frame = +3
Query: 254 VGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFW 313
VGHPLEPYLSLLKLDGKLILMGVI TPLQFITPMVMLGRRSITGSFIGSIKETEEM+EFW
Sbjct: 3 VGHPLEPYLSLLKLDGKLILMGVIYTPLQFITPMVMLGRRSITGSFIGSIKETEEMVEFW 182
Query: 314 KEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKLIDQ 358
KEKGLTSMIEIVNMDYI+K FERLEKNDVRYRFVVDV GS+LIDQ
Sbjct: 183 KEKGLTSMIEIVNMDYIHKPFERLEKNDVRYRFVVDV*GSQLIDQ 317
>AV764937
Length = 392
Score = 119 bits (299), Expect = 6e-28
Identities = 61/129 (47%), Positives = 86/129 (66%), Gaps = 1/129 (0%)
Frame = +1
Query: 139 IIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKES-GLRGGILGLGGVGHMGV 197
++ + ++VV IPE + + APLLCA +TV+SPL L S G + G++GLGG+GH+ V
Sbjct: 4 MVADYRYVVHIPENLPMDAAAPLLCAGITVFSPLKDHDLVSSPGKKIGVVGLGGLGHIAV 183
Query: 198 IIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHP 257
KA GHHVTVIS+S K+ EA + LGAD +++S++ +Q A SLD++ DTV H
Sbjct: 184 KFGKAFGHHVTVISTSPSKEAEAKQRLGADDFILSTNQQHLQSARRSLDFVSDTVSAEHS 363
Query: 258 LEPYLSLLK 266
L P L LLK
Sbjct: 364 LLPLLELLK 390
>BP056571
Length = 319
Score = 68.9 bits (167), Expect = 1e-12
Identities = 31/60 (51%), Positives = 44/60 (72%)
Frame = -1
Query: 291 GRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
GR++I S IG IKET+EM++F + + IE++ MDY+N A ERL K DV+YRFV+D+
Sbjct: 292 GRKTIAXSMIGGIKETQEMIDFAAKHDVKPEIEVIPMDYVNTAMERLLKADVKYRFVIDI 113
>TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, partial (60%)
Length = 724
Score = 64.3 bits (155), Expect = 3e-11
Identities = 55/206 (26%), Positives = 94/206 (44%), Gaps = 5/206 (2%)
Frame = +1
Query: 27 FTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNY----PMVPGHEVVGEVLEVGSDV 82
F L GP DV +++ G+C +DVH +K L +++ PMV GHE G + VG++V
Sbjct: 139 FKLPTLGPHDVRVRMKAVGICGSDVHYLKK-LRCADFIVKEPMVIGHECAGIIEAVGAEV 315
Query: 83 TRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVE 142
G+ V + C +C C+ C + + P G A I+
Sbjct: 316 KSLVPGDRVAIEPGISCW-RCDHCKLGSYNLCP------DMKFFATPPVHGSLANQIVHP 474
Query: 143 QKFVVKIPEGMAPEQVAPLLCAAVTVYS-PLSHFGLKESGLRGGILGLGGVGHMGVIIAK 201
K+P+ ++ E+ A +V V++ ++ G + + L I+G G +G + ++ A+
Sbjct: 475 ADLCFKLPDNVSLEEGAMCEPLSVGVHACRRANIGPETNVL---IMGAGPIGLVTMLAAR 645
Query: 202 AMGHHVTVISSSDRKKKEAIEDLGAD 227
A G VI D + + LGAD
Sbjct: 646 AFGAPRIVIVDVDDHRLSVAKTLGAD 723
>TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , complete
Length = 1489
Score = 63.5 bits (153), Expect = 5e-11
Identities = 86/366 (23%), Positives = 143/366 (38%), Gaps = 32/366 (8%)
Frame = +1
Query: 8 RTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVP 67
R V W A P ++ G +V +K+ Y +CHTDV+ + +P +
Sbjct: 145 RAAVSWEAGKPL-VIEEVEVAPPQAG--EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIF 315
Query: 68 GHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYC-------KKKIW- 119
GHE G V VG VT G+ + G C +C C+S+ C + +
Sbjct: 316 GHEAGGIVESVGEGVTHLKPGD-HALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMI 492
Query: 120 --NYNDVYVDGKPT-----QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPL 172
N + + GKP F+E ++ V KI ++V L C T +
Sbjct: 493 SDNQSRFSIKGKPIYHFVGTSTFSEYTVLHAGCVAKINPAAPLDKVCILSCGICTGFGAT 672
Query: 173 SHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVS 232
+ + G I GLG VG A+ G + + E + G + ++
Sbjct: 673 VNVAKPKPGSSVAIFGLGAVGLAAAEGARVSGASRIIGVDLVSSRFEGAKKFGVNEFVNP 852
Query: 233 SD-TTSMQEA-ADSLDYIID-TVPVGHPLEPYLSLLKLD----GKLILMGVINTPLQFIT 285
D +QE A+ + +D V ++ +S + G +L+GV N F T
Sbjct: 853 KDHDKPVQEVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNQDDAFQT 1032
Query: 286 -PMVMLGRRSITGSFIGSIKETEEML---------EFWKEKGLTSMIEIVNMDYINKAFE 335
P+ L R++ G+F G+ K ++ E EK +T V+ IN+AF+
Sbjct: 1033HPVNFLNERTLKGTFYGNYKPRTDLPNVVEMYMRGELELEKFIT---HTVSFSEINQAFD 1203
Query: 336 RLEKND 341
+ K +
Sbjct: 1204YMLKGE 1221
>TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, partial (98%)
Length = 1908
Score = 56.2 bits (134), Expect = 8e-09
Identities = 52/197 (26%), Positives = 84/197 (42%), Gaps = 5/197 (2%)
Frame = +2
Query: 61 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN 120
S P PG++V G V++VGS+V +F VG+ V +
Sbjct: 905 SPLPTAPGYDVAGVVVKVGSEVKKFKVGDEVYGDI------------------------- 1009
Query: 121 YNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKES 180
N+ ++ G AE VE+K + P+ ++ + A L A T Y GL+ +
Sbjct: 1010 -NENALEYPKVVGSLAEYTAVEEKLLAHKPKNLSFVEAASLPLAIETAYE-----GLERA 1171
Query: 181 GLRGG--ILGLGGVGHMG---VIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDT 235
G G IL LGG G +G + +AK + V ++S K E + LGAD +
Sbjct: 1172 GFSAGKSILVLGGAGGVGTHVIQLAKHVFGASKVAATSSTGKLELLRKLGAD-MPIDYTK 1348
Query: 236 TSMQEAADSLDYIIDTV 252
+++ + D + D V
Sbjct: 1349 ENVENLPEKFDVVFDAV 1399
>BP039342
Length = 535
Score = 48.9 bits (115), Expect = 1e-06
Identities = 22/33 (66%), Positives = 23/33 (69%)
Frame = -3
Query: 103 CTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGF 135
C ACQ DIEQY KK WNYN V+VD KP G F
Sbjct: 167 CRACQYDIEQYRNKKNWNYNGVFVDRKPHCGVF 69
>BP048139
Length = 567
Score = 48.1 bits (113), Expect = 2e-06
Identities = 38/143 (26%), Positives = 55/143 (37%), Gaps = 15/143 (10%)
Frame = -2
Query: 63 YPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKK------ 116
+P + GHE VG V VG DVT T G++V + + C +CT C+S C
Sbjct: 431 FPRILGHEAVGVVESVGKDVTEVTKGDVV-IPIYLPDCGECTDCKSTKSNRCTNFPFKVS 255
Query: 117 ---------KIWNYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVT 167
+ + N + F+E +V+ V KI + P++ L C T
Sbjct: 254 PSMPRDGTTRFTDLNGEIIHHFMFISSFSEYTVVDVANVTKIDPEIPPDRACLLSCGIST 75
Query: 168 VYSPLSHFGLKESGLRGGILGLG 190
E G I GLG
Sbjct: 74 GVGAAWRTASVEPGSTVAIFGLG 6
>AV422928
Length = 399
Score = 46.6 bits (109), Expect = 6e-06
Identities = 20/31 (64%), Positives = 25/31 (80%)
Frame = -3
Query: 323 EIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
E++ D IN+A ERL KNDVRYRFV+D+ GS
Sbjct: 397 ELITPDRINEAMERLAKNDVRYRFVIDIAGS 305
>AV423166
Length = 452
Score = 46.2 bits (108), Expect = 8e-06
Identities = 25/81 (30%), Positives = 40/81 (48%)
Frame = +2
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V +K+ +CHTD+ + + +P+ GHE VG V VG VT G+++
Sbjct: 158 EVRVKMLCASICHTDITSI-HGFPHGIFPLALGHEGVGIVESVGDQVTNLKEGDMI-IPT 331
Query: 96 LVGCCKKCTACQSDIEQYCKK 116
+G C++C C S C K
Sbjct: 332 YIGECQECENCVSGETNLCMK 394
>TC18928 similar to UP|Q43677 (Q43677) Auxin-induced protein, partial (53%)
Length = 612
Score = 44.7 bits (104), Expect = 2e-05
Identities = 46/166 (27%), Positives = 70/166 (41%), Gaps = 4/166 (2%)
Frame = +1
Query: 35 DDVYIKVHYCGVCHTDVHQVKNDLGMSNYPM--VPGHEVVGEVLEVGSDVTRFTVGEIVG 92
D V IKV + D +++ S+ P+ PG++V G V++VGS+V +F VG+ V
Sbjct: 175 DQVLIKVAAASLNPIDYKRMEGAFKASDSPLPTAPGYDVAGVVVKVGSEVKKFKVGDEV- 351
Query: 93 AGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEG 152
DI + VY G AE E+K + P+
Sbjct: 352 --------------YGDINEQAM--------VY---PKVVGSLAEYTAAEEKLLSHKPQN 456
Query: 153 MAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGG--ILGLGGVGHMG 196
++ + A L T Y+ GL+ +G G IL LGG G +G
Sbjct: 457 LSFAEAASLPLTIETAYN-----GLELAGFSAGKSILVLGGAGGVG 579
>AW719877
Length = 520
Score = 37.4 bits (85), Expect = 0.004
Identities = 34/128 (26%), Positives = 60/128 (46%), Gaps = 7/128 (5%)
Frame = +2
Query: 122 NDVYVDGK-PTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPL-SHFGLKE 179
+DV+ G QG AE +V+++ V + P + Q A L A+T + L +K+
Sbjct: 110 DDVFYSGALAPQGTNAEFHLVDERLVGRKPSSLDYAQAAALPLTAITAFEALFDRLDVKK 289
Query: 180 S--GLRGGIL---GLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSD 234
G IL G GGVG + V +A+ + + ++S + ++ + LGA + S
Sbjct: 290 PVVGAANAILIIGGAGGVGSIAVQLARQLTDLTVIATASRPETRDWVLGLGAHHVIDHSK 469
Query: 235 TTSMQEAA 242
+ + AA
Sbjct: 470 PLAAEVAA 493
>TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , partial
(33%)
Length = 694
Score = 37.4 bits (85), Expect = 0.004
Identities = 23/79 (29%), Positives = 36/79 (45%)
Frame = +3
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V +K+ Y +CHTDV+ + +P + GHE G V VG VT + +
Sbjct: 387 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPVD-HALPV 563
Query: 96 LVGCCKKCTACQSDIEQYC 114
G C + C+S+ C
Sbjct: 564 FTGECGELPHCKSEESNMC 620
>TC9889 similar to UP|Q9AYU1 (Q9AYU1) Quinone-oxidoreductase QR1
(Fragment), partial (49%)
Length = 528
Score = 35.0 bits (79), Expect = 0.019
Identities = 38/138 (27%), Positives = 51/138 (36%), Gaps = 6/138 (4%)
Frame = +1
Query: 62 NYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNY 121
++P +P +V GEV EVG V F VG+ V A L QY
Sbjct: 202 SFPHIPCTDVAGEVAEVGPQVKDFKVGDKVIAKLST--------------QY-------- 315
Query: 122 NDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFG---LK 178
GG AE + + P ++P + A L A +T L+ L
Sbjct: 316 ----------GGGLAEFAVASETLTAARPSQVSPAEAAGLPIAGLTARDALTQIAGVKLH 465
Query: 179 ESGLRGGIL---GLGGVG 193
E+G IL GGVG
Sbjct: 466 ETGQPKNILVTAASGGVG 519
>BI420064
Length = 410
Score = 33.1 bits (74), Expect = 0.074
Identities = 12/29 (41%), Positives = 20/29 (68%)
Frame = +3
Query: 63 YPMVPGHEVVGEVLEVGSDVTRFTVGEIV 91
+P +PG ++ GEV+E+G V +F G+ V
Sbjct: 288 FPYIPGTDIAGEVVEIGQGVKKFKPGDKV 374
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,761,871
Number of Sequences: 28460
Number of extensions: 74556
Number of successful extensions: 379
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 372
length of query: 358
length of database: 4,897,600
effective HSP length: 91
effective length of query: 267
effective length of database: 2,307,740
effective search space: 616166580
effective search space used: 616166580
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0290.23


