

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.14
(461 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
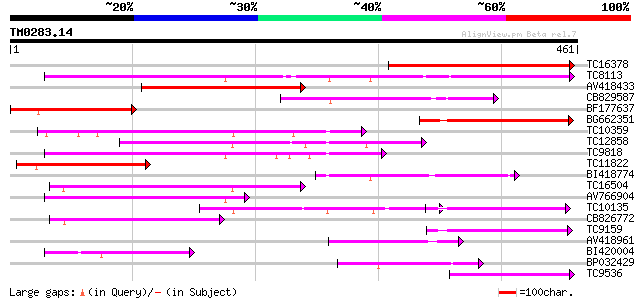
Score E
Sequences producing significant alignments: (bits) Value
TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22... 218 2e-57
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 159 7e-40
AV418433 142 2e-34
CB829587 118 2e-27
BF177637 118 2e-27
BG662351 110 4e-25
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 109 1e-24
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 108 2e-24
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 107 5e-24
TC11822 similar to UP|P93051 (P93051) ORFc protein (Fragment), p... 88 3e-18
BI418774 87 4e-18
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 85 3e-17
AV766904 83 1e-16
TC10135 75 2e-14
CB826772 73 1e-13
TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose tr... 72 1e-13
AV418961 70 6e-13
BI420004 64 4e-11
BP032429 62 3e-10
TC9536 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, parti... 61 3e-10
>TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;}, partial (31%)
Length = 738
Score = 218 bits (555), Expect = 2e-57
Identities = 105/151 (69%), Positives = 130/151 (85%)
Frame = +2
Query: 309 ATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALI 368
+TWLVDKSGRR+LL++SSSIMT+SL++V+ AFYLEG VS+ S L+ ILGI+SVVGLV ++
Sbjct: 2 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 181
Query: 369 IGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTI 428
IGF+LG+GPIPW+IMSEILP +IKGLAGS AT +NW T+ IT+TAN LL WSS GTFT+
Sbjct: 182 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 361
Query: 429 YAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
Y + +AFTV F L+VPETK RTLEEIQ+SF
Sbjct: 362 YTVVAAFTVVFTALWVPETKGRTLEEIQSSF 454
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 159 bits (403), Expect = 7e-40
Identities = 123/472 (26%), Positives = 212/472 (44%), Gaps = 41/472 (8%)
Frame = +2
Query: 29 FVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSG 88
+ + CV+V ++ I G+ G S + D+ +S ++ + + N+ A+VG +G
Sbjct: 113 YALACVIVASMVSIISGYDTGVMSGALLFIKEDIGISDTQQEVLAGILNICALVGCLAAG 292
Query: 89 HLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEI 148
++Y GR+ ++ +A++ + G V + + ++L GR + G GVG PVY AE+
Sbjct: 293 KTSDYIGRRYTIFLASILFLVGAVFMGYGPNFAILMFGRCVCGLGVGFALTTAPVYSAEL 472
Query: 149 PPRTMRGSLGAVNQLSVTIGIMLAYL-------LGLFFNWRTLALLGVFPCAILIPGLYF 201
+ RG L ++ ++ + +GI + Y+ L L WR + L P L G+
Sbjct: 473 SSASTRGFLTSLPEVCIGLGIFIGYISNYFLGKLALTLGWRLMLGLAAIPSLGLALGILT 652
Query: 202 IPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFYTVCRPQEEKILVSF--- 258
+PESPRWL G + + L L SN T E E++ R V +EK F
Sbjct: 653 MPESPRWLVMQGRLGCAKKVL--LEVSN---TTEEAELRFRDIIVSAGFDEKCNDEFVKQ 817
Query: 259 ---------------------------DGIGLLVLQQLSGINGVFFYASKIFASAGISSS 291
+G+ + +GI V Y +IF AG+++
Sbjct: 818 PQKSHHGEGVWKELFLRPTPPVRRMLIAAVGIHFFEHATGIEAVMLYGPRIFKKAGVTTK 997
Query: 292 D---AATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSK 348
D AT G G ++ ++T+L+D+ GRR LL +S + M L ++ F L V
Sbjct: 998 DRLLLATIGTGLTKITFLTISTFLLDRVGRRRLLQISVAGMIFGLTIL--GFSLTMVEYS 1171
Query: 349 GSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTAS 408
+L L LS+V + F +G+ P+ W+ SEI P ++ S+ N +
Sbjct: 1172SEKLVWALS-LSIVATYTYVAFFNVGLAPVTWVYSSEIFPLRLRAQGNSIGVAVNRGMNA 1348
Query: 409 VITLT-ANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
I+++ + + G F ++A S F +PETK + LEE++ F
Sbjct: 1349AISMSFISIYKAITIGGAFFLFAGMSVVAWVFFYFCLPETKGKALEEMEMVF 1504
>AV418433
Length = 401
Score = 142 bits (357), Expect = 2e-34
Identities = 65/133 (48%), Positives = 96/133 (71%)
Frame = +3
Query: 108 IFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTI 167
+ GW+ I ++ LL IGR+ G+G+G+ SYVVPV+IAEI P+ +RG+L +NQ +
Sbjct: 3 VAGWLVIYFSQGPVLLDIGRVATGYGMGVFSYVVPVFIAEIAPKELRGALTTLNQFMLVS 182
Query: 168 GIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRG 227
+ +++++G +WR LAL G+ P A+L+ GL+FIPESPRWLA+ G E FE++LQ LRG
Sbjct: 183 AMSVSFVIGNVLSWRALALTGLIPTAVLLLGLFFIPESPRWLAKRGCREDFEAALQILRG 362
Query: 228 SNVDITMEAQEIQ 240
+ DI+ EA+EIQ
Sbjct: 363 KDADISQEAEEIQ 401
>CB829587
Length = 548
Score = 118 bits (295), Expect = 2e-27
Identities = 72/186 (38%), Positives = 108/186 (57%), Gaps = 9/186 (4%)
Frame = +2
Query: 221 SLQTLRGSNVDITMEAQEIQNRFYTVCRPQEEKILVSFD---------GIGLLVLQQLSG 271
+L+ LRG +VDI+ EA EI + T+ + K+L F G+GL+V QQ G
Sbjct: 2 ALRRLRGKDVDISHEATEILDSIETLQSLPKSKLLDLFHTRYVRSVVIGVGLMVCQQSVG 181
Query: 272 INGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTI 331
ING+ FY ++ F +AG+SS A T IQV T + L+DKSGRR L+ VSS +
Sbjct: 182 INGIGFYTAETFVAAGVSSGKAGTIAYACIQVPFTILGAILMDKSGRRPLITVSSGGTFL 361
Query: 332 SLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNI 391
+ AF+L+ +G L + L++ G++ I +++G+G +PW+IMSE+ P +I
Sbjct: 362 GCFITGIAFFLK---DQGLLLEWV-PTLAIGGVLIYIAAYSIGLGSVPWVIMSEVFPIHI 529
Query: 392 KGLAGS 397
KG AGS
Sbjct: 530 KGTAGS 547
>BF177637
Length = 480
Score = 118 bits (295), Expect = 2e-27
Identities = 62/122 (50%), Positives = 78/122 (63%), Gaps = 19/122 (15%)
Frame = +2
Query: 1 MPLGEDYEDSRNIRKPFINNNN-------------------AGSGSFFVVLCVLVVALGP 41
M E+ E+ R+++KPF++ + S V CVL+VALGP
Sbjct: 113 MSYREENEEGRDLKKPFLHTGSWYRMSGKQQSNLFASAHEAIRDSSISVFACVLIVALGP 292
Query: 42 IQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLL 101
IQFGFT GY+SPTQ +I DL LS+S FSLFGSLSNVGAM+GA SG +AEY GRKGSL+
Sbjct: 293 IQFGFTAGYTSPTQAAIISDLGLSVSEFSLFGSLSNVGAMLGAIASGQIAEYIGRKGSLM 472
Query: 102 VA 103
+A
Sbjct: 473 IA 478
>BG662351
Length = 407
Score = 110 bits (276), Expect = 4e-25
Identities = 56/126 (44%), Positives = 86/126 (67%), Gaps = 1/126 (0%)
Frame = +1
Query: 334 LLVASAFYLEGV-VSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIK 392
+L A AFYL+ +S G+ + L+V G++ + FA+G+G +PW++MSEI P NIK
Sbjct: 22 ILTAVAFYLKAQEISVGA-----VPGLAVTGILVYVGSFAIGMGAVPWVVMSEIFPVNIK 186
Query: 393 GLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTL 452
G AGS+AT NWF A + + T NFL++WS+ GTF +YA +A + ++ VPETK ++L
Sbjct: 187 GQAGSLATLVNWFGAWLCSYTFNFLMSWSTYGTFILYAAINALGILSIVVVVPETKGKSL 366
Query: 453 EEIQAS 458
E++QA+
Sbjct: 367 EQLQAA 384
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 109 bits (272), Expect = 1e-24
Identities = 82/312 (26%), Positives = 149/312 (47%), Gaps = 44/312 (14%)
Frame = +2
Query: 23 AGSGSF------FVVLCVLVVALGPIQFGFTCGYSSPT----------------QEEMIR 60
+G G F V++ ++ A G + FG+ G S + + +
Sbjct: 170 SGGGDFEAKITPIVIISCILAATGGLMFGYDVGVSGGVTSMPHFLHKFFPDVYKKTVLEK 349
Query: 61 DLNLSISRFS-----LFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAIS 115
+L+ + ++ LF S + +V + + GR+ ++L+A V I G V
Sbjct: 350 ELDSNYCKYDNQGLQLFTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFFIAGVVFNV 529
Query: 116 LAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLL 175
A++ ++L +GR+L G GVG + VP++++EI P +RG+L + QL++TIGI+ A L+
Sbjct: 530 TAQNLAMLIVGRILLGCGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITIGILFANLI 709
Query: 176 GLFFN-------WRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGS 228
N WR L P +L G + ++P L E G E ++ L+ +RG+
Sbjct: 710 NYGTNKIKGGWGWRLSLGLAGIPAVLLTVGALLVVDTPNSLIERGRTEEGKAVLKKIRGT 889
Query: 229 N---------VDITMEAQEIQNRFYTVC-RPQEEKILVSFDGIGLLVLQQLSGINGVFFY 278
+ V+ + A+E+++ F + R +I++S I L + QQ +GIN + FY
Sbjct: 890 DNVEPEFLELVEASRVAKEVKDPFRNLLKRRNRPQIIIS---IALQIFQQFTGINAIMFY 1060
Query: 279 ASKIFASAGISS 290
A +F + G +
Sbjct: 1061APVLFNTLGFKN 1096
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 108 bits (269), Expect = 2e-24
Identities = 78/269 (28%), Positives = 131/269 (47%), Gaps = 19/269 (7%)
Frame = +3
Query: 90 LAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIP 149
+ G + +++V + + G + LA +L +GRLL GFG+G + VP+Y++E+
Sbjct: 21 ITRIMGMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIYLSEMA 200
Query: 150 PRTMRGSLGAVNQLSVTIGIMLAYLLGLFF-------NWRTLALLGVFPCAILIPGLYFI 202
P RG L QLS+TIGI +A L +F WR LG P I + G +
Sbjct: 201 PYKYRGGLNMCFQLSITIGIFVANLFNYYFAKILNGQGWRLSLGLGAVPAVIFVVGSLCL 380
Query: 203 PESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEI----------QNRFYTVCRPQEE 252
P+SP L G E L +RG+ DI E ++I ++ + T+ +
Sbjct: 381 PDSPSSLVARGRHEAARQELVKIRGT-TDIEAELKDIITASEALENVKHPWKTLLERKYR 557
Query: 253 KILVSFDGIGLLVLQQLSGINGVFFYASKIFASAGI--SSSDAATFGLGAIQVVMTGVAT 310
LV + + QQ +G+N + FYA +F + G ++S + +G+ + V T ++
Sbjct: 558 PQLVF--AVCIPFFQQFTGLNVITFYAPILFRTIGFGPTASLMSAVIIGSFKPVSTLISI 731
Query: 311 WLVDKSGRRILLIVSSSIMTISLLLVASA 339
++VDK GRR L + + M I +++ A
Sbjct: 732 FVVDKFGRRTLFLEGGAQMLICQIIMTIA 818
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 107 bits (266), Expect = 5e-24
Identities = 77/311 (24%), Positives = 136/311 (42%), Gaps = 33/311 (10%)
Frame = +2
Query: 29 FVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSG 88
+ C ++ ++ I G+ G S + RDL +S + + + N+ +++G+ ++G
Sbjct: 233 YAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDVKIEILLGIINLYSLIGSGLAG 412
Query: 89 HLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEI 148
+++ GR+ +++ A G + + + + L GR + G G+G + PVY AE+
Sbjct: 413 RTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYWFLMFGRFIAGIGIGYALMIAPVYTAEV 592
Query: 149 PPRTMRGSLGAVNQLSVTIGIMLAYL-------LGLFFNWRTLALLGVFPCAILIPGLYF 201
P + RG L + ++ + GI+L Y+ L L WR + +G P IL G+
Sbjct: 593 SPASSRGFLTSFPEVFINGGILLGYISNFAFSKLSLKVGWRMMLGVGALPSVILGVGVLA 772
Query: 202 IPESPRWLAEMGLM--------------ERFESSLQTLR---GSNVDITMEAQEIQNR-- 242
+PESPRWL G + E + L ++ G T + E+ R
Sbjct: 773 MPESPRWLVMRGRLGDAIKVLNKTSDSPEEAQLRLADIKRAAGIPESCTDDVVEVSKRST 952
Query: 243 -------FYTVCRPQEEKILVSFDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAAT 295
+ P I+++ +G+ QQ SGI+ V Y+ IF AGI S
Sbjct: 953 GEGVWKELFLYPTPAIRHIVIA--ALGIHFFQQASGIDAVVLYSPTIFEKAGIKSDTDKL 1126
Query: 296 FGLGAIQVVMT 306
A+ V T
Sbjct: 1127LATVAVGFVKT 1159
>TC11822 similar to UP|P93051 (P93051) ORFc protein (Fragment), partial
(74%)
Length = 419
Score = 87.8 bits (216), Expect = 3e-18
Identities = 46/115 (40%), Positives = 73/115 (63%), Gaps = 6/115 (5%)
Frame = +3
Query: 6 DYEDSRNIRKPFINN-----NNAGSGSFFVVLCVLVVAL-GPIQFGFTCGYSSPTQEEMI 59
+Y + IR+P + + N+A G ++V VA+ G +FG GYSSPTQ+ +
Sbjct: 57 EYGVEKGIREPLVKDQNKLLNHARKGHPWIVYFTTFVAVCGSYEFGAGIGYSSPTQDAIR 236
Query: 60 RDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAI 114
+DLNLS++ +SLFGS+ GAM+GA SG +A++ GRKG++ V++ + GW+ I
Sbjct: 237 KDLNLSLAEYSLFGSIVTFGAMIGAITSGLIADFIGRKGAMRVSSAFCVAGWLVI 401
>BI418774
Length = 554
Score = 87.4 bits (215), Expect = 4e-18
Identities = 56/170 (32%), Positives = 94/170 (54%), Gaps = 4/170 (2%)
Frame = +3
Query: 249 PQEEKILVSFDGIGLLVLQQLSGINGVFFYASKIFASAGISSSD---AATFGLGAIQVVM 305
P +IL++ IG+ V QQ+ GI G+ Y+ ++F GI+S AT GLG Q V
Sbjct: 54 PPVRRILIA--AIGVHVFQQICGIEGICIYSPRVFEKTGITSKSNLLLATVGLGISQAVF 227
Query: 306 TGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLV 365
T ++ +L+D+ GRRILL++SS + ++LL ++ + + + S+ + GI + +
Sbjct: 228 TFISAFLLDRVGRRILLLISSGGVVVTLLGLSFCSF----IMQQSKEEPLWGISFTIVAI 395
Query: 366 ALIIGF-ALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTA 414
+ + F A+GIGP+ W+ SEI P ++ V N A+V LT+
Sbjct: 396 YIFVAFVAIGIGPVTWVYSSEIFPLRLRAQGLGVCVLVNRL-ANVAVLTS 542
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 84.7 bits (208), Expect = 3e-17
Identities = 58/217 (26%), Positives = 96/217 (43%), Gaps = 9/217 (4%)
Frame = +2
Query: 33 CVLVVALGPI--QFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHL 90
C VVA I +G+ + + DL LS + L ++ A+ +G +
Sbjct: 89 CASVVAASVIFALYGYVMAVMTGVLLFIKEDLQLSDLQVQFSVGLLHMSALPACMTAGRI 268
Query: 91 AEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPP 150
A+Y GR+ ++++ A+ + G V + +L IG + G G+G V PVY AEI P
Sbjct: 269 ADYIGRRYTIILTAIIFLVGSVLMGYGPSYPILMIGLSISGIGLGFSLIVAPVYCAEISP 448
Query: 151 RTMRGSLGAVNQLSVTIGIMLAYLLGLFF-------NWRTLALLGVFPCAILIPGLYFIP 203
+ RG L + ++S GI L Y+ FF WR + ++ P L+ +
Sbjct: 449 PSHRGFLTSFLEVSTNAGIALGYVSNYFFGKLSIRLGWRLMLVVHAVPSLALVALMLNWV 628
Query: 204 ESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQ 240
ESPRWL G + L + + + +EI+
Sbjct: 629 ESPRWLVMQGRVGEARKVLLLVSNTKEEAEQRLKEIK 739
>AV766904
Length = 611
Score = 82.8 bits (203), Expect = 1e-16
Identities = 42/174 (24%), Positives = 87/174 (49%), Gaps = 7/174 (4%)
Frame = +3
Query: 29 FVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSG 88
F + C ++ ++ I G+ G S + RDL +S + + + N+ + +G+ ++G
Sbjct: 87 FALACAILASMTSILLGYDIGVMSGAVIYIKRDLKISDVQIEILSGIMNIYSPIGSYIAG 266
Query: 89 HLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEI 148
+++ GR+ ++++A + G + + + + + L GR + G G+G + PVY +EI
Sbjct: 267 RTSDWIGRRYTIVLAGLIFFVGAILMGFSPNYAFLMFGRFVAGVGIGFAFLIAPVYTSEI 446
Query: 149 PPRTMRGSLGAVNQLSVTIGIMLAYL-------LGLFFNWRTLALLGVFPCAIL 195
P RG L ++ ++ + GI++ Y+ L L WR + +G P L
Sbjct: 447 SPTLSRGFLTSLPEVFLNAGILIGYISNYTFSKLPLRLGWRLMLGIGAIPSIFL 608
>TC10135
Length = 1501
Score = 75.1 bits (183), Expect = 2e-14
Identities = 66/223 (29%), Positives = 106/223 (46%), Gaps = 24/223 (10%)
Frame = +2
Query: 155 GSLGAVNQLSVTIGIMLAYLLGLFFN-----WRTLALLGVFPCAILIPGLYFIPESPRWL 209
G+L ++N +T G L+YL+ L F WR + + P I I + +PESPRWL
Sbjct: 2 GALVSLNSFLITGGQFLSYLINLAFTNAPGTWRWMLGVAAAPAIIQIVLMLSLPESPRWL 181
Query: 210 AEMGLMERFESSLQTLRG-SNVDITMEAQEIQNRFYTVCRPQEEKILVS----------- 257
G E +S L+ + +VD +EA + ++ + + KI +
Sbjct: 182 YRKGREEESKSILKKIYAPEDVDAEIEALK-ESVESEIEESKTSKISMMTLLKTTTVRRG 358
Query: 258 -FDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAA------TFGLGAIQVVMTGVAT 310
+ G+GL QQ GIN V +Y+ I AG +S+ A T GL A ++ +
Sbjct: 359 LYAGMGLQFFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLITSGLNAFGSIL---SI 529
Query: 311 WLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELH 353
+ +DK+GR+ L ++S + +SL L+ AF + SELH
Sbjct: 530 YFIDKTGRKKLALISLCGVVLSLALLTVAF-------RESELH 637
Score = 70.5 bits (171), Expect = 6e-13
Identities = 38/119 (31%), Positives = 66/119 (54%), Gaps = 1/119 (0%)
Frame = +2
Query: 339 AFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSV 398
A+Y G SK G ++VGL II F+ G+G +PW+I SEI P +G+ G +
Sbjct: 851 AWYTRGCPSK-------FGWAALVGLALYIITFSPGMGTVPWVINSEIYPFRYRGVCGGI 1009
Query: 399 ATFSNWFTASVITLT-ANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQ 456
A+ + W + +++ + + +A TF ++ I + + F ++FVPETK +EE++
Sbjct: 1010ASTTVWISNLIVSQSFLSLTQTIGTAWTFMMFGILAVVAIFFVIVFVPETKGVPMEEVE 1186
>CB826772
Length = 490
Score = 72.8 bits (177), Expect = 1e-13
Identities = 38/144 (26%), Positives = 75/144 (51%), Gaps = 2/144 (1%)
Frame = +1
Query: 33 CVLVVALGPIQ--FGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHL 90
CV V+A + +G+ G + + DL +S + + ++ + + +G +
Sbjct: 58 CVSVMAASILSAIYGYEVGVMTGALLFIKEDLQISDLQLQFLVGIFHMCGLPASMAAGRI 237
Query: 91 AEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPP 150
++Y GR+ ++++A++ + G + + +L IG + G G G V P+Y AEI P
Sbjct: 238 SDYIGRRYTIILASIAFLSGSILTGYSPSYLILMIGNFISGIGAGFTLIVAPLYCAEISP 417
Query: 151 RTMRGSLGAVNQLSVTIGIMLAYL 174
+ RGSL ++ + S+ IGI+L Y+
Sbjct: 418 PSHRGSLTSLTEFSINIGILLGYM 489
>TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(5%)
Length = 728
Score = 72.4 bits (176), Expect = 1e-13
Identities = 39/119 (32%), Positives = 68/119 (56%), Gaps = 1/119 (0%)
Frame = +3
Query: 340 FYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVA 399
+Y +G SK LG L++ GL II F+ G+G +PW++ SEI P +G+ G +A
Sbjct: 81 WYTQGCPSK-------LGFLALGGLALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIA 239
Query: 400 TFSNWFTASVITLT-ANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQA 457
+ + W + V++ + + +A TF ++ I + + F L+FVPETK +EE+++
Sbjct: 240 STTVWVSNLVVSQSFLSLTQTIGTAWTFMMFGIIAIIGIFFVLIFVPETKGVPMEEVES 416
>AV418961
Length = 394
Score = 70.5 bits (171), Expect = 6e-13
Identities = 44/110 (40%), Positives = 65/110 (59%)
Frame = +3
Query: 260 GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRR 319
G L + QQL+GIN V +Y++ +F SAGI+S AA+ +GA V T VA+ L+D+ GR+
Sbjct: 81 GAALFLFQQLAGINAVVYYSTSVFRSAGIASDVAASALVGASNVFGTAVASSLMDRQGRK 260
Query: 320 ILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALII 369
LLI S S M S+LL++ +F L G L+V+G V ++
Sbjct: 261 SLLITSFSGMAASMLLLSLSF-------TWKVLTPYSGTLAVLGTVLYVL 389
>BI420004
Length = 547
Score = 64.3 bits (155), Expect = 4e-11
Identities = 42/126 (33%), Positives = 64/126 (50%), Gaps = 4/126 (3%)
Frame = +1
Query: 29 FVVLCVLVVALGPIQFGFTCGYSSPTQEEMIRDLNLSISRFSLFG----SLSNVGAMVGA 84
+++ +G + FG+ G S I+D + R SL S++ VGA++GA
Sbjct: 166 YILGVTAAAGIGGLLFGYDTGVISGALL-YIKDDFEEVRRSSLLQETIVSMALVGAIIGA 342
Query: 85 TVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVY 144
G + + FGRK + L A V G + ++ A D+ + GRLL G G+G+ S PVY
Sbjct: 343 ATGGWINDAFGRKKATLSADVVFTLGSLVMAAAPDAYFVIFGRLLVGLGIGVASVTAPVY 522
Query: 145 IAEIPP 150
IAE P
Sbjct: 523 IAESSP 540
>BP032429
Length = 468
Score = 61.6 bits (148), Expect = 3e-10
Identities = 42/122 (34%), Positives = 63/122 (51%), Gaps = 3/122 (2%)
Frame = -2
Query: 267 QQLSGINGVFFYASKIFASAGISSSDAATFGL--GAIQVVMTGVATWLVDKSGRRILLIV 324
QQL+GIN + FYA +F + G A + G + V+ T V+ + VDK GRR+L +
Sbjct: 371 QQLTGINVIMFYAPVLFKTLGFGDDAALMSAVISGGVNVLATFVSIFSVDKFGRRLLFLE 192
Query: 325 SSSIMTISLLLVASAFYLE-GVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIM 383
M IS + V + L+ GV +GS +L + + A + FA GP+ WL+
Sbjct: 191 GGVQMLISQIAVGTMIALKFGVSGEGSFTKGEANLL-LFFICAYVAAFAWSWGPLGWLVP 15
Query: 384 SE 385
SE
Sbjct: 14 SE 9
>TC9536 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (17%)
Length = 537
Score = 61.2 bits (147), Expect = 3e-10
Identities = 36/103 (34%), Positives = 51/103 (48%), Gaps = 1/103 (0%)
Frame = +2
Query: 358 ILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFL 417
I+S V +V F + GPIP ++ SEI P ++GL ++ W ++T T +
Sbjct: 20 IISTVCVVVYFCFFVMAYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIVTYTLPVM 199
Query: 418 L-NWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
L + AG F +YAI + F L VPETK LE I F
Sbjct: 200 LSSMGLAGVFGLYAIVCCISWVFVYLKVPETKGMPLEVITEFF 328
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.141 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,485,182
Number of Sequences: 28460
Number of extensions: 98641
Number of successful extensions: 652
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 623
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 629
length of query: 461
length of database: 4,897,600
effective HSP length: 93
effective length of query: 368
effective length of database: 2,250,820
effective search space: 828301760
effective search space used: 828301760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0283.14


