

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.13
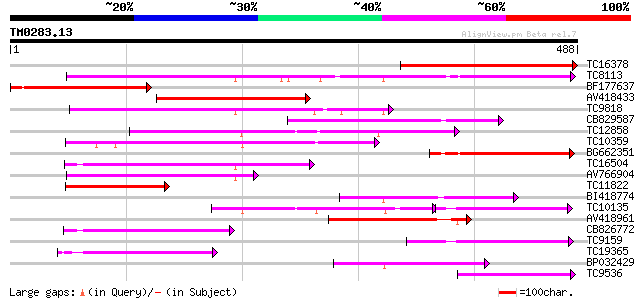
(488 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22... 295 9e-81
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 189 1e-48
BF177637 180 4e-46
AV418433 147 5e-36
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 134 3e-32
CB829587 130 6e-31
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 127 4e-30
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 114 4e-26
BG662351 112 1e-25
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 97 6e-21
AV766904 97 6e-21
TC11822 similar to UP|P93051 (P93051) ORFc protein (Fragment), p... 97 6e-21
BI418774 96 2e-20
TC10135 89 2e-18
AV418961 86 1e-17
CB826772 84 7e-17
TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose tr... 79 1e-15
TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter l... 66 1e-11
BP032429 66 1e-11
TC9536 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, parti... 66 1e-11
>TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;}, partial (31%)
Length = 738
Score = 295 bits (756), Expect = 9e-81
Identities = 152/152 (100%), Positives = 152/152 (100%)
Frame = +2
Query: 337 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 396
STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV
Sbjct: 2 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMV 181
Query: 397 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 456
IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM
Sbjct: 182 IGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTM 361
Query: 457 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 488
YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR
Sbjct: 362 YTVVAAFTVVFTALWVPETKGRTLEEIQSSFR 457
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 189 bits (479), Expect = 1e-48
Identities = 135/469 (28%), Positives = 236/469 (49%), Gaps = 31/469 (6%)
Frame = +2
Query: 50 LFCVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIASGQI 109
L CV++A++ I G+ G S I D+ +S ++ + + N+ A+VG +A+G+
Sbjct: 119 LACVIVASMVSIISGYDTGVMSGALLFIKEDIGISDTQQEVLAGILNICALVGCLAAGKT 298
Query: 110 AEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVYIAEIAP 169
++Y+GR+ ++ +A+I ++G + + + +++ L GR + G GVG PVY AE++
Sbjct: 299 SDYIGRRYTIFLASILFLVGAVFMGYGPNFAILMFGRCVCGLGVGFALTTAPVYSAELSS 478
Query: 170 ENLRGGLGSVNQLSVTIGILLAYL-------LGLFVNWRVLAILGILPCTILIPGLFFIP 222
+ RG L S+ ++ + +GI + Y+ L L + WR++ L +P L G+ +P
Sbjct: 479 ASTRGFLTSLPEVCIGLGIFIGYISNYFLGKLALTLGWRLMLGLAAIPSLGLALGILTMP 658
Query: 223 ESPRWLAKMG--------------MTEEFE---SSLQVLRGFDTDISVE-VHEIKRSIVP 264
ESPRWL G TEE E + V GFD + E V + ++S
Sbjct: 659 ESPRWLVMQGRLGCAKKVLLEVSNTTEEAELRFRDIIVSAGFDEKCNDEFVKQPQKSHHG 838
Query: 265 RG--RRVAIRFADLQRKRYWFPLMVGIGLLVLQQLSGINGVLFYSTSIFANAGISSSS-- 320
G + + +R R+ L+ +G+ + +GI V+ Y IF AG+++
Sbjct: 839 EGVWKELFLRPTPPVRRM----LIAAVGIHFFEHATGIEAVMLYGPRIFKKAGVTTKDRL 1006
Query: 321 -AATVGLGAIQVIATGISTWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSH 379
AT+G G ++ IST+L+D+ GRR LL +S + M L ++ + + SE
Sbjct: 1007LLATIGTGLTKITFLTISTFLLDRVGRRRLLQISVAGMIFGLTILGFSLTMVEYSSE--K 1180
Query: 380 LYKILGIVSVVGLVVMVIGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAIT 439
L L + S+V V F++GL P+ WV SEI P+ ++ S N + AI+
Sbjct: 1181LVWALSL-SIVATYTYVAFFNVGLAPVTWVYSSEIFPLRLRAQGNSIGVAVNRGMNAAIS 1357
Query: 440 MT-ANLLLTWSSGGTFTMYTVVAAFTVVFTALWVPETKGRTLEEIQSSF 487
M+ ++ + GG F ++ ++ VF +PETKG+ LEE++ F
Sbjct: 1358MSFISIYKAITIGGAFFLFAGMSVVAWVFFYFCLPETKGKALEEMEMVF 1504
>BF177637
Length = 480
Score = 180 bits (457), Expect = 4e-46
Identities = 92/123 (74%), Positives = 107/123 (86%), Gaps = 1/123 (0%)
Frame = +2
Query: 1 MSFREESGEGRGDLQKPFLHTGSWYKM-GSRQSSVMGSTTQVMRDGSVSVLFCVLIAALG 59
MS+REE+ EGR DL+KPFLHTGSWY+M G +QS++ S + +RD S+SV CVLI ALG
Sbjct: 113 MSYREENEEGR-DLKKPFLHTGSWYRMSGKQQSNLFASAHEAIRDSSISVFACVLIVALG 289
Query: 60 PIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIASGQIAEYVGRKGSL 119
PIQFGFT GY+SPTQ AI+SDL LS+SEFSLFGSLSNVGAM+GAIASGQIAEY+GRKGSL
Sbjct: 290 PIQFGFTAGYTSPTQAAIISDLGLSVSEFSLFGSLSNVGAMLGAIASGQIAEYIGRKGSL 469
Query: 120 MIA 122
MIA
Sbjct: 470 MIA 478
>AV418433
Length = 401
Score = 147 bits (370), Expect = 5e-36
Identities = 67/133 (50%), Positives = 98/133 (73%)
Frame = +3
Query: 127 IIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVYIAEIAPENLRGGLGSVNQLSVTI 186
+ GWL I F++ L +GR+ G+G+G+ SYVVPV+IAEIAP+ LRG L ++NQ +
Sbjct: 3 VAGWLVIYFSQGPVLLDIGRVATGYGMGVFSYVVPVFIAEIAPKELRGALTTLNQFMLVS 182
Query: 187 GILLAYLLGLFVNWRVLAILGILPCTILIPGLFFIPESPRWLAKMGMTEEFESSLQVLRG 246
+ +++++G ++WR LA+ G++P +L+ GLFFIPESPRWLAK G E+FE++LQ+LRG
Sbjct: 183 AMSVSFVIGNVLSWRALALTGLIPTAVLLLGLFFIPESPRWLAKRGCREDFEAALQILRG 362
Query: 247 FDTDISVEVHEIK 259
D DIS E EI+
Sbjct: 363 KDADISQEAEEIQ 401
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 134 bits (337), Expect = 3e-32
Identities = 88/306 (28%), Positives = 155/306 (49%), Gaps = 27/306 (8%)
Frame = +2
Query: 52 CVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIASGQIAE 111
C ++A++ I G+ G S I DLK+S + + + N+ +++G+ +G+ ++
Sbjct: 245 CAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDVKIEILLGIINLYSLIGSGLAGRTSD 424
Query: 112 YVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVYIAEIAPEN 171
++GR+ +++ A +G L + F+ +Y FL GR + G G+G + PVY AE++P +
Sbjct: 425 WIGRRYTIVFAGAIFFVGALLMGFSPNYWFLMFGRFIAGIGIGYALMIAPVYTAEVSPAS 604
Query: 172 LRGGLGSVNQLSVTIGILLAYL-------LGLFVNWRVLAILGILPCTILIPGLFFIPES 224
RG L S ++ + GILL Y+ L L V WR++ +G LP IL G+ +PES
Sbjct: 605 SRGFLTSFPEVFINGGILLGYISNFAFSKLSLKVGWRMMLGVGALPSVILGVGVLAMPES 784
Query: 225 PRWLAKMGMTEEFESSLQVLRGFDTDISVEVHEIKRS----------IVPRGRRVAIRFA 274
PRWL G + L + + + +IKR+ +V +R
Sbjct: 785 PRWLVMRGRLGDAIKVLNKTSDSPEEAQLRLADIKRAAGIPESCTDDVVEVSKRST--GE 958
Query: 275 DLQRKRYWFP-------LMVGIGLLVLQQLSGINGVLFYSTSIFANAGISSSS---AATV 324
+ ++ + +P ++ +G+ QQ SGI+ V+ YS +IF AGI S + ATV
Sbjct: 959 GVWKELFLYPTPAIRHIVIAALGIHFFQQASGIDAVVLYSPTIFEKAGIKSDTDKLLATV 1138
Query: 325 GLGAIQ 330
+G ++
Sbjct: 1139AVGFVK 1156
>CB829587
Length = 548
Score = 130 bits (326), Expect = 6e-31
Identities = 74/186 (39%), Positives = 109/186 (57%)
Frame = +2
Query: 240 SLQVLRGFDTDISVEVHEIKRSIVPRGRRVAIRFADLQRKRYWFPLMVGIGLLVLQQLSG 299
+L+ LRG D DIS E EI SI + DL RY +++G+GL+V QQ G
Sbjct: 2 ALRRLRGKDVDISHEATEILDSIETLQSLPKSKLLDLFHTRYVRSVVIGVGLMVCQQSVG 181
Query: 300 INGVLFYSTSIFANAGISSSSAATVGLGAIQVIATGISTWLVDKSGRRLLLLLSSSIMTV 359
ING+ FY+ F AG+SS A T+ IQV T + L+DKSGRR L+ +SS +
Sbjct: 182 INGIGFYTAETFVAAGVSSGKAGTIAYACIQVPFTILGAILMDKSGRRPLITVSSGGTFL 361
Query: 360 SLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMVIGFSLGLGPIPWVIMSEILPVSI 419
+ IAF+L+ + L + + +++ G+++ + +S+GLG +PWVIMSE+ P+ I
Sbjct: 362 GCFITGIAFFLK----DQGLLLEWVPTLAIGGVLIYIAAYSIGLGSVPWVIMSEVFPIHI 529
Query: 420 KGLAGS 425
KG AGS
Sbjct: 530 KGTAGS 547
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 127 bits (319), Expect = 4e-30
Identities = 88/294 (29%), Positives = 146/294 (48%), Gaps = 10/294 (3%)
Frame = +3
Query: 104 IASGQIAEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVY 163
+ + I +G + ++++ + + G L A L +GRLL GFG+G + VP+Y
Sbjct: 6 LGASTITRIMGMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIY 185
Query: 164 IAEIAPENLRGGLGSVNQLSVTIGILLAYLLGLFV-------NWRVLAILGILPCTILIP 216
++E+AP RGGL QLS+TIGI +A L + WR+ LG +P I +
Sbjct: 186 LSEMAPYKYRGGLNMCFQLSITIGIFVANLFNYYFAKILNGQGWRLSLGLGAVPAVIFVV 365
Query: 217 GLFFIPESPRWLAKMGMTEEFESSLQVLRGFDTDISVEVHEIKRSIVPRGRRVAIRFADL 276
G +P+SP L G E L +RG TDI E+ +I + V + L
Sbjct: 366 GSLCLPDSPSSLVARGRHEAARQELVKIRG-TTDIEAELKDIITASEAL-ENVKHPWKTL 539
Query: 277 QRKRYWFPLMVGIGLLVLQQLSGINGVLFYSTSIFANAGI--SSSSAATVGLGAIQVIAT 334
++Y L+ + + QQ +G+N + FY+ +F G ++S + V +G+ + ++T
Sbjct: 540 LERKYRPQLVFAVCIPFFQQFTGLNVITFYAPILFRTIGFGPTASLMSAVIIGSFKPVST 719
Query: 335 GISTWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLE-GTVSEDSHLYKILGIV 387
IS ++VDK GRR L L + M + ++++IA + GT L K +V
Sbjct: 720 LISIFVVDKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTSGNPGQLPKWYAVV 881
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 114 bits (285), Expect = 4e-26
Identities = 80/298 (26%), Positives = 144/298 (47%), Gaps = 28/298 (9%)
Frame = +2
Query: 49 VLFCVLIAALGPIQFGFTCGYSSPT----------------QDAIVSDLKLSLSEFS--- 89
V+ ++AA G + FG+ G S + + +L + ++
Sbjct: 209 VIISCILAATGGLMFGYDVGVSGGVTSMPHFLHKFFPDVYKKTVLEKELDSNYCKYDNQG 388
Query: 90 --LFGSLSNVGAMVGAIASGQIAEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRL 147
LF S + +V + +GR+ +++IA + I G + A++ + L +GR+
Sbjct: 389 LQLFTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFFIAGVVFNVTAQNLAMLIVGRI 568
Query: 148 LEGFGVGIISYVVPVYIAEIAPENLRGGLGSVNQLSVTIGILLAYLLGLFVN-------W 200
L G GVG + VP++++EIAP +RG L + QL++TIGIL A L+ N W
Sbjct: 569 LLGCGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITIGILFANLINYGTNKIKGGWGW 748
Query: 201 RVLAILGILPCTILIPGLFFIPESPRWLAKMGMTEEFESSLQVLRGFDTDISVEVHEIKR 260
R+ L +P +L G + ++P L + G TEE ++ L+ +RG D + ++
Sbjct: 749 RLSLGLAGIPAVLLTVGALLVVDTPNSLIERGRTEEGKAVLKKIRGTDNVEPEFLELVEA 928
Query: 261 SIVPRGRRVAIRFADLQRKRYWFPLMVGIGLLVLQQLSGINGVLFYSTSIFANAGISS 318
S V + V F +L ++R +++ I L + QQ +GIN ++FY+ +F G +
Sbjct: 929 SRV--AKEVKDPFRNLLKRRNRPQIIISIALQIFQQFTGINAIMFYAPVLFNTLGFKN 1096
>BG662351
Length = 407
Score = 112 bits (281), Expect = 1e-25
Identities = 51/125 (40%), Positives = 88/125 (69%)
Frame = +1
Query: 362 VVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMVIGFSLGLGPIPWVIMSEILPVSIKG 421
++ ++AFYL+ +++ + + G+ +V G++V V F++G+G +PWV+MSEI PV+IKG
Sbjct: 22 ILTAVAFYLK---AQEISVGAVPGL-AVTGILVYVGSFAIGMGAVPWVVMSEIFPVNIKG 189
Query: 422 LAGSTATMANWLTSWAITMTANLLLTWSSGGTFTMYTVVAAFTVVFTALWVPETKGRTLE 481
AGS AT+ NW +W + T N L++WS+ GTF +Y + A ++ + VPETKG++LE
Sbjct: 190 QAGSLATLVNWFGAWLCSYTFNFLMSWSTYGTFILYAAINALGILSIVVVVPETKGKSLE 369
Query: 482 EIQSS 486
++Q++
Sbjct: 370 QLQAA 384
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 97.1 bits (240), Expect = 6e-21
Identities = 62/222 (27%), Positives = 110/222 (48%), Gaps = 7/222 (3%)
Frame = +2
Query: 48 SVLFCVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIASG 107
SV+ +I AL +G+ + I DL+LS + L ++ A+ + +G
Sbjct: 95 SVVAASVIFAL----YGYVMAVMTGVLLFIKEDLQLSDLQVQFSVGLLHMSALPACMTAG 262
Query: 108 QIAEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVYIAEI 167
+IA+Y+GR+ ++++ AI ++G + + + Y L +G + G G+G V PVY AEI
Sbjct: 263 RIADYIGRRYTIILTAIIFLVGSVLMGYGPSYPILMIGLSISGIGLGFSLIVAPVYCAEI 442
Query: 168 APENLRGGLGSVNQLSVTIGILLAYL-------LGLFVNWRVLAILGILPCTILIPGLFF 220
+P + RG L S ++S GI L Y+ L + + WR++ ++ +P L+ +
Sbjct: 443 SPPSHRGFLTSFLEVSTNAGIALGYVSNYFFGKLSIRLGWRLMLVVHAVPSLALVALMLN 622
Query: 221 IPESPRWLAKMGMTEEFESSLQVLRGFDTDISVEVHEIKRSI 262
ESPRWL G E L ++ + + EIK ++
Sbjct: 623 WVESPRWLVMQGRVGEARKVLLLVSNTKEEAEQRLKEIKVAV 748
>AV766904
Length = 611
Score = 97.1 bits (240), Expect = 6e-21
Identities = 49/172 (28%), Positives = 96/172 (55%), Gaps = 7/172 (4%)
Frame = +3
Query: 50 LFCVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIASGQI 109
L C ++A++ I G+ G S I DLK+S + + + N+ + +G+ +G+
Sbjct: 93 LACAILASMTSILLGYDIGVMSGAVIYIKRDLKISDVQIEILSGIMNIYSPIGSYIAGRT 272
Query: 110 AEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVYIAEIAP 169
++++GR+ ++++A + +G + + F+ +Y+FL GR + G G+G + PVY +EI+P
Sbjct: 273 SDWIGRRYTIVLAGLIFFVGAILMGFSPNYAFLMFGRFVAGVGIGFAFLIAPVYTSEISP 452
Query: 170 ENLRGGLGSVNQLSVTIGILLAYL-------LGLFVNWRVLAILGILPCTIL 214
RG L S+ ++ + GIL+ Y+ L L + WR++ +G +P L
Sbjct: 453 TLSRGFLTSLPEVFLNAGILIGYISNYTFSKLPLRLGWRLMLGIGAIPSIFL 608
>TC11822 similar to UP|P93051 (P93051) ORFc protein (Fragment), partial
(74%)
Length = 419
Score = 97.1 bits (240), Expect = 6e-21
Identities = 46/89 (51%), Positives = 63/89 (70%)
Frame = +3
Query: 49 VLFCVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIASGQ 108
V F +A G +FG GYSSPTQDAI DL LSL+E+SLFGS+ GAM+GAI SG
Sbjct: 147 VYFTTFVAVCGSYEFGAGIGYSSPTQDAIRKDLNLSLAEYSLFGSIVTFGAMIGAITSGL 326
Query: 109 IAEYVGRKGSLMIAAIPNIIGWLAISFAK 137
IA+++GRKG++ +++ + GWL I F++
Sbjct: 327 IADFIGRKGAMRVSSAFCVAGWLVIYFSQ 413
>BI418774
Length = 554
Score = 95.5 bits (236), Expect = 2e-20
Identities = 56/158 (35%), Positives = 90/158 (56%), Gaps = 4/158 (2%)
Frame = +3
Query: 285 LMVGIGLLVLQQLSGINGVLFYSTSIFANAGISSSS---AATVGLGAIQVIATGISTWLV 341
L+ IG+ V QQ+ GI G+ YS +F GI+S S ATVGLG Q + T IS +L+
Sbjct: 72 LIAAIGVHVFQQICGIEGICIYSPRVFEKTGITSKSNLLLATVGLGISQAVFTFISAFLL 251
Query: 342 DKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGI-VSVVGLVVMVIGFS 400
D+ GRR+LLL+SS + V+L+ +S + + + S + GI ++V + + V +
Sbjct: 252 DRVGRRILLLISSGGVVVTLLGLSFCSF----IMQQSKEEPLWGISFTIVAIYIFVAFVA 419
Query: 401 LGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAI 438
+G+GP+ WV SEI P+ ++ + N L + A+
Sbjct: 420 IGIGPVTWVYSSEIFPLRLRAQGLGVCVLVNRLANVAV 533
>TC10135
Length = 1501
Score = 89.0 bits (219), Expect = 2e-18
Identities = 69/210 (32%), Positives = 106/210 (49%), Gaps = 15/210 (7%)
Frame = +2
Query: 174 GGLGSVNQLSVTIGILLAYLLGL-FVN----WRVLAILGILPCTILIPGLFFIPESPRWL 228
G L S+N +T G L+YL+ L F N WR + + P I I + +PESPRWL
Sbjct: 2 GALVSLNSFLITGGQFLSYLINLAFTNAPGTWRWMLGVAAAPAIIQIVLMLSLPESPRWL 181
Query: 229 AKMGMTEEFESSLQVLRGFDTDISVEVHEIKRSI---VPRGRRVAIRFAD-LQRKRYWFP 284
+ G EE +S L+ + D+ E+ +K S+ + + I L+
Sbjct: 182 YRKGREEESKSILKKIYA-PEDVDAEIEALKESVESEIEESKTSKISMMTLLKTTTVRRG 358
Query: 285 LMVGIGLLVLQQLSGINGVLFYSTSIFANAGISSSSAA------TVGLGAIQVIATGIST 338
L G+GL QQ GIN V++YS +I AG +S+ A T GL A I +S
Sbjct: 359 LYAGMGLQFFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLITSGLNAFGSI---LSI 529
Query: 339 WLVDKSGRRLLLLLSSSIMTVSLVVVSIAF 368
+ +DK+GR+ L L+S + +SL ++++AF
Sbjct: 530 YFIDKTGRKKLALISLCGVVLSLALLTVAF 619
Score = 76.6 bits (187), Expect = 8e-15
Identities = 40/119 (33%), Positives = 68/119 (56%), Gaps = 1/119 (0%)
Frame = +2
Query: 367 AFYLEGTVSEDSHLYKILGIVSVVGLVVMVIGFSLGLGPIPWVIMSEILPVSIKGLAGST 426
A+Y G S+ G ++VGL + +I FS G+G +PWVI SEI P +G+ G
Sbjct: 851 AWYTRGCPSK-------FGWAALVGLALYIITFSPGMGTVPWVINSEIYPFRYRGVCGGI 1009
Query: 427 ATMANWLTSWAITMT-ANLLLTWSSGGTFTMYTVVAAFTVVFTALWVPETKGRTLEEIQ 484
A+ W+++ ++ + +L T + TF M+ ++A + F ++VPETKG +EE++
Sbjct: 1010ASTTVWISNLIVSQSFLSLTQTIGTAWTFMMFGILAVVAIFFVIVFVPETKGVPMEEVE 1186
>AV418961
Length = 394
Score = 86.3 bits (212), Expect = 1e-17
Identities = 49/127 (38%), Positives = 79/127 (61%), Gaps = 4/127 (3%)
Frame = +3
Query: 275 DLQRKRYWFPLMVGIGLLVLQQLSGINGVLFYSTSIFANAGISSSSAATVGLGAIQVIAT 334
DL RYW + VG L + QQL+GIN V++YSTS+F +AGI+S AA+ +GA V T
Sbjct: 42 DLFSTRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIASDVAASALVGASNVFGT 221
Query: 335 GISTWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKIL----GIVSVV 390
+++ L+D+ GR+ LL+ S S M S++++S++F +K+L G ++V+
Sbjct: 222 AVASSLMDRQGRKSLLITSFSGMAASMLLLSLSF-----------TWKVLTPYSGTLAVL 368
Query: 391 GLVVMVI 397
G V+ V+
Sbjct: 369 GTVLYVL 389
>CB826772
Length = 490
Score = 83.6 bits (205), Expect = 7e-17
Identities = 43/147 (29%), Positives = 84/147 (56%)
Frame = +1
Query: 47 VSVLFCVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMVGAIAS 106
VSV+ +++A+ +G+ G + I DL++S + + ++ + ++A+
Sbjct: 61 VSVMAASILSAI----YGYEVGVMTGALLFIKEDLQISDLQLQFLVGIFHMCGLPASMAA 228
Query: 107 GQIAEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVPVYIAE 166
G+I++Y+GR+ ++++A+I + G + ++ Y L +G + G G G V P+Y AE
Sbjct: 229 GRISDYIGRRYTIILASIAFLSGSILTGYSPSYLILMIGNFISGIGAGFTLIVAPLYCAE 408
Query: 167 IAPENLRGGLGSVNQLSVTIGILLAYL 193
I+P + RG L S+ + S+ IGILL Y+
Sbjct: 409 ISPPSHRGSLTSLTEFSINIGILLGYM 489
>TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(5%)
Length = 728
Score = 79.3 bits (194), Expect = 1e-15
Identities = 45/145 (31%), Positives = 78/145 (53%), Gaps = 1/145 (0%)
Frame = +3
Query: 342 DKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVMVIGFSL 401
DK LL + T V +Y +G S+ LG +++ GL + +I FS
Sbjct: 3 DKLSPGACLLSDDNTRTQCQKVHHRPWYTQGCPSK-------LGFLALGGLALYIIFFSP 161
Query: 402 GLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMT-ANLLLTWSSGGTFTMYTVV 460
G+G +PWV+ SEI P+ +G+ G A+ W+++ ++ + +L T + TF M+ ++
Sbjct: 162 GMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSLTQTIGTAWTFMMFGII 341
Query: 461 AAFTVVFTALWVPETKGRTLEEIQS 485
A + F ++VPETKG +EE++S
Sbjct: 342 AIIGIFFVLIFVPETKGVPMEEVES 416
>TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (24%)
Length = 441
Score = 66.2 bits (160), Expect = 1e-11
Identities = 38/138 (27%), Positives = 75/138 (53%)
Frame = +2
Query: 42 MRDGSVSVLFCVLIAALGPIQFGFTCGYSSPTQDAIVSDLKLSLSEFSLFGSLSNVGAMV 101
+RDG +S+L+ + FG+ G + I +L ++ + L G + NV A+
Sbjct: 50 LRDG-LSLLYLAI--------FGYVTGVMAGALLFIKEELGINDMQVQLLGGILNVCALP 202
Query: 102 GAIASGQIAEYVGRKGSLMIAAIPNIIGWLAISFAKDYSFLYMGRLLEGFGVGIISYVVP 161
+ +G+ ++YVGR+ +++++A+ ++G + + + + L +GR G GVG+ + P
Sbjct: 203 ACMVAGRTSDYVGRRYTIILSAVILLLGSILMGYGPSFPILMIGRCTAGLGVGLALIIAP 382
Query: 162 VYIAEIAPENLRGGLGSV 179
VY EI+ + RG L S+
Sbjct: 383 VYSTEISLPSNRGFLTSL 436
>BP032429
Length = 468
Score = 65.9 bits (159), Expect = 1e-11
Identities = 45/138 (32%), Positives = 69/138 (49%), Gaps = 3/138 (2%)
Frame = -2
Query: 279 KRYWFPLMVGIGLL-VLQQLSGINGVLFYSTSIFANAGISSSSA--ATVGLGAIQVIATG 335
K + P MV L+ QQL+GIN ++FY+ +F G +A + V G + V+AT
Sbjct: 422 KPVYKPQMVFCSLIPFFQQLTGINVIMFYAPVLFKTLGFGDDAALMSAVISGGVNVLATF 243
Query: 336 ISTWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYKILGIVSVVGLVVM 395
+S + VDK GRRLL L M +S + V L+ VS + K + + +
Sbjct: 242 VSIFSVDKFGRRLLFLEGGVQMLISQIAVGTMIALKFGVSGEGSFTKGEANLLLFFICAY 63
Query: 396 VIGFSLGLGPIPWVIMSE 413
V F+ GP+ W++ SE
Sbjct: 62 VAAFAWSWGPLGWLVPSE 9
>TC9536 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (17%)
Length = 537
Score = 65.9 bits (159), Expect = 1e-11
Identities = 37/103 (35%), Positives = 54/103 (51%), Gaps = 1/103 (0%)
Frame = +2
Query: 386 IVSVVGLVVMVIGFSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLL 445
I+S V +VV F + GPIP ++ SEI P ++GL + + W+ +T T ++
Sbjct: 20 IISTVCVVVYFCFFVMAYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIVTYTLPVM 199
Query: 446 L-TWSSGGTFTMYTVVAAFTVVFTALWVPETKGRTLEEIQSSF 487
L + G F +Y +V + VF L VPETKG LE I F
Sbjct: 200 LSSMGLAGVFGLYAIVCCISWVFVYLKVPETKGMPLEVITEFF 328
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.139 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,358,739
Number of Sequences: 28460
Number of extensions: 113082
Number of successful extensions: 636
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 600
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 607
length of query: 488
length of database: 4,897,600
effective HSP length: 94
effective length of query: 394
effective length of database: 2,222,360
effective search space: 875609840
effective search space used: 875609840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0283.13


