

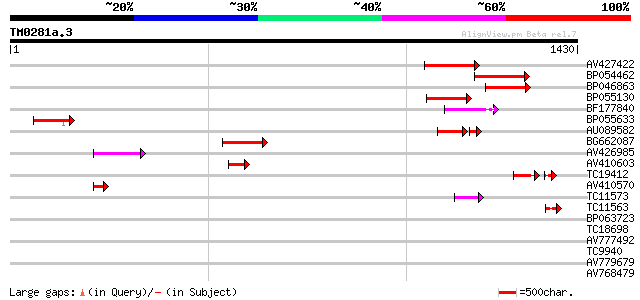
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.3
(1430 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV427422 214 6e-56
BP054462 171 1e-42
BP046863 137 9e-33
BP055130 133 2e-31
BF177840 99 6e-21
BP055633 98 8e-21
AU089582 78 2e-19
BG662087 91 1e-18
AV426985 86 3e-17
AV410603 72 5e-13
TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%) 59 2e-09
AV410570 49 6e-06
TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase regi... 49 7e-06
TC11563 42 7e-04
BP063723 39 0.006
TC18698 39 0.008
AV777492 32 0.92
TC9940 weakly similar to UP|Q9LHB1 (Q9LHB1) Transcription factor... 31 1.2
AV779679 31 1.6
AV768479 31 1.6
>AV427422
Length = 417
Score = 214 bits (546), Expect = 6e-56
Identities = 91/137 (66%), Positives = 117/137 (84%)
Frame = +1
Query: 1047 GLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDP 1106
GLPK +G + VLVVVDRL+KF+HF L HPY+AK +A +F++E+V+LHG P +IVSDRDP
Sbjct: 7 GLPKSKGYEAVLVVVDRLSKFSHFVPLKHPYTAKVIADIFVREVVRLHGVPLSIVSDRDP 186
Query: 1107 LFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPW 1166
LF+SNFWKELFKM GT+LK+ST+YHP++DGQTEV NRCLETYLRCF+ +PK W W+PW
Sbjct: 187 LFMSNFWKELFKMQGTKLKMSTAYHPESDGQTEVVNRCLETYLRCFIADQPKSWAHWVPW 366
Query: 1167 AEFWFNTNYNSSAGMSP 1183
AE+W+NT+Y+ S G +P
Sbjct: 367 AEYWYNTSYHVSTGQTP 417
>BP054462
Length = 422
Score = 171 bits (432), Expect = 1e-42
Identities = 81/140 (57%), Positives = 109/140 (77%)
Frame = +1
Query: 1172 NTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQH 1231
N+NYN SA MSPF+ALYGR+PP+LL+ IPS++ VN L RD +L +L+ NLLK+Q
Sbjct: 1 NSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAVNDLQVGRDELLSDLRANLLKSQD 180
Query: 1232 QMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAY 1291
M+ A+K RR V++ GD V+LKLQPY+ +S+A + KL+PR+YGPY I+ ++G VAY
Sbjct: 181 MMRTYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAY 360
Query: 1292 KLELPAHSKIHPVFHVSLLK 1311
+LELPAHS++HPVFHVSLLK
Sbjct: 361 RLELPAHSRVHPVFHVSLLK 420
>BP046863
Length = 580
Score = 137 bits (346), Expect = 9e-33
Identities = 65/113 (57%), Positives = 90/113 (79%)
Frame = +1
Query: 1200 AIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPY 1259
+I VE VN L ++RD +L EL+ NLLKAQ QM+ QA+KHRR V++ G+WV+LKLQPY
Sbjct: 241 SIKLSVEAVNMLIEERDALLLELRGNLLKAQDQMRAQANKHRRYVDYQVGNWVFLKLQPY 420
Query: 1260 KLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFHVSLLKQ 1312
KL+++A R KL+PRFYGP+K+L+RV VAY L+L + S++HPVFH+SLL++
Sbjct: 421 KLQNLAQRKNQKLSPRFYGPFKVLERVVQVAY*LDLXSESRVHPVFHLSLLEK 579
>BP055130
Length = 567
Score = 133 bits (334), Expect = 2e-31
Identities = 62/114 (54%), Positives = 77/114 (67%)
Frame = +2
Query: 1051 VRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLS 1110
V G ++VVVDRL+K+ HF Y++ VA F+ +VKLHG P IVSDRD F S
Sbjct: 173 VCGFTVIIVVVDRLSKYGHFAPHRANYTSSQVAETFVSTIVKLHGMPRAIVSDRDKAFTS 352
Query: 1111 NFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWL 1164
FWK FK+ GT L +S+SYHPQTDGQTE N+CLE YLRCFV P+ W+ +L
Sbjct: 353 AFWKHFFKLHGTTLNMSSSYHPQTDGQTEALNKCLELYLRCFVHETPRLWVSYL 514
>BF177840
Length = 410
Score = 98.6 bits (244), Expect = 6e-21
Identities = 55/135 (40%), Positives = 77/135 (56%)
Frame = +2
Query: 1098 TTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKP 1157
T+IVSDRD F+S+FW+ L+ GT+L ST+ HPQTDGQTEV N+ L T LR +
Sbjct: 11 TSIVSDRDTKFISHFWRTLWGKVGTKLLYSTTCHPQTDGQTEVVNKTLSTLLRSVLERNL 190
Query: 1158 KQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDG 1217
K W WLP EF +N +S+ SPF+ +YG +P L +P+ L + +D
Sbjct: 191 KMWETWLPHIEFAYNRVVHSTTKHSPFEIVYGYNPLTPLDLLPMPN-----TYLLKHKD- 352
Query: 1218 VLGELKQNLLKAQHQ 1232
G+ K +K H+
Sbjct: 353 --GKAKAEFVKRLHE 391
>BP055633
Length = 528
Score = 98.2 bits (243), Expect = 8e-21
Identities = 52/113 (46%), Positives = 78/113 (69%), Gaps = 10/113 (8%)
Frame = -2
Query: 61 TWEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVRGIF 120
TW FK A++ +FQ + +PF LL+LKQ G+V+E+V +FE++AG LK ID + IF
Sbjct: 521 TWTKFKEAMLEQFQLTSNSSPFAALLALKQEGSVEEFVGQFERFAGMLKGIDEEHYMDIF 342
Query: 121 LNGLKEEIKAEVRL---------IQKSLLIEQKNQIVTKKGVGFT-RPSSAFR 163
+NGLKEEI AE++L ++K+L++EQKN V+K G+G T R +S+F+
Sbjct: 341 VNGLKEEIAAEIKLYEPKSLTIMVKKALMVEQKNLAVSKAGIGSTSRYNSSFK 183
>AU089582
Length = 383
Score = 77.8 bits (190), Expect(2) = 2e-19
Identities = 36/76 (47%), Positives = 51/76 (66%)
Frame = +2
Query: 1078 SAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQ 1137
+A A +++ E+V LHG P +I+SDR F S+FW+ GT+LK+ST++HPQTDGQ
Sbjct: 8 AASQYAKIYLDEIVSLHGVPVSIISDRGAQFTSHFWRSFQTALGTRLKMSTAFHPQTDGQ 187
Query: 1138 TEVTNRCLETYLRCFV 1153
+E T + LE LR V
Sbjct: 188 SERTIQILEDMLRACV 235
Score = 36.6 bits (83), Expect(2) = 2e-19
Identities = 16/31 (51%), Positives = 21/31 (67%)
Frame = +3
Query: 1160 WLDWLPWAEFWFNTNYNSSAGMSPFKALYGR 1190
W +L EF +N +Y SS M+PF+ALYGR
Sbjct: 255 WDQYLSLMEFAYNNSYRSSI*MAPFEALYGR 347
>BG662087
Length = 373
Score = 91.3 bits (225), Expect = 1e-18
Identities = 45/113 (39%), Positives = 70/113 (61%)
Frame = +1
Query: 537 GGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEK 596
G WR+ VDY LNK + +P+P I++L+D ++ S +D SGYHQI+M +D +K
Sbjct: 16 GKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDEDK 195
Query: 597 TAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYS 649
TAF T +Y + +PFGL NA +T+Q LM+ V + + V+ D+++V S
Sbjct: 196 TAFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKS 354
>AV426985
Length = 422
Score = 86.3 bits (212), Expect = 3e-17
Identities = 46/135 (34%), Positives = 75/135 (55%), Gaps = 5/135 (3%)
Frame = +1
Query: 212 KHLTGAQMREKRDKNLCFRCDEPYSREHKCRNKQLRMIIMEDDDDADCGEFEDALS---- 267
K+++ A+M+ +R+KNLC+ CDE +S HKC NK L M+ + D++D+D + +
Sbjct: 10 KYISPAEMQLRREKNLCYWCDEKFSFNHKCPNKYLMMLQLTDENDSDDTSNQQVTTTISD 189
Query: 268 -GEFNSLQLSLCSMAGLTSTKSWKMVGQIGNQSIVVLIDCGASHNFISHKLVEQLNLTVA 326
SL +M G T + + GQIGN S+ VL+D G+S F+ + + L L +
Sbjct: 190 DSPVEDHHFSLNAMRGFTGVGTIRFTGQIGNISVQVLVDGGSSDCFLQPHIAKFLQLPIE 369
Query: 327 ETQTYTVEVGDGYKI 341
+ V VG+G K+
Sbjct: 370 SKPNFKVLVGNGQKM 414
>AV410603
Length = 162
Score = 72.4 bits (176), Expect = 5e-13
Identities = 32/53 (60%), Positives = 41/53 (76%)
Frame = +1
Query: 552 TIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEG 604
T+ + FP+P ++ELLDEL G++ FSKLDL+SGYHQI +K D KT FRTH G
Sbjct: 4 TVKDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRTHHG 162
>TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%)
Length = 519
Score = 59.3 bits (142), Expect(2) = 2e-09
Identities = 30/66 (45%), Positives = 47/66 (70%), Gaps = 1/66 (1%)
Frame = +3
Query: 1271 KLAPRFYGPYKILDRVGPVAYKLELPAH-SKIHPVFHVSLLKQALKPTQQPQPLPPMLNE 1329
KL+PRF GP+++L+RVG V+Y+L LP S +HPVFHVS+L++ L P + +E
Sbjct: 48 KLSPRFIGPFEVLERVGSVSYRLALPPDLSAVHPVFHVSMLRKYLY-----DPSHVIRHE 212
Query: 1330 ELELEV 1335
+++L+V
Sbjct: 213 DVQLDV 230
Score = 20.8 bits (42), Expect(2) = 2e-09
Identities = 9/30 (30%), Positives = 18/30 (60%)
Frame = +2
Query: 1349 GNLEVLIKWEQLPTCDNTWELAAVIQDKFP 1378
G+++VL W + TWE ++++K+P
Sbjct: 305 GSVKVL--WRGPSGEEATWEAEDIMREKYP 388
>AV410570
Length = 412
Score = 48.9 bits (115), Expect = 6e-06
Identities = 20/37 (54%), Positives = 27/37 (72%)
Frame = +2
Query: 212 KHLTGAQMREKRDKNLCFRCDEPYSREHKCRNKQLRM 248
K ++ A+M+ +R+K LCF CDE YS HKC NKQL +
Sbjct: 83 KRISPAEMQLRREKGLCFTCDEKYSWNHKCPNKQLML 193
>TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase region
(Fragment), partial (10%)
Length = 572
Score = 48.5 bits (114), Expect = 7e-06
Identities = 26/74 (35%), Positives = 40/74 (53%)
Frame = +2
Query: 1121 GTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAG 1180
G Q++ S+ HPQT+GQTE N+ + ++ + +W+D LP + +NT SS
Sbjct: 62 GIQMRFSSVKHPQTNGQTEAANKVILKGIKRRLYEAEGRWIDELPIVLWSYNTMPQSSIK 241
Query: 1181 MSPFKALYGRDPPL 1194
+PF YG D L
Sbjct: 242 ETPF*LTYGADTML 283
>TC11563
Length = 470
Score = 42.0 bits (97), Expect = 7e-04
Identities = 20/39 (51%), Positives = 24/39 (61%)
Frame = +2
Query: 1352 EVLIKWEQLPTCDNTWELAAVIQDKFPLFPLEDKVKLLG 1390
++LI+WE C TWEL + IQD FP F L DKV G
Sbjct: 14 QLLIQWEGAANC--TWELLSYIQDSFPQFALADKVTFYG 124
>BP063723
Length = 479
Score = 38.9 bits (89), Expect = 0.006
Identities = 18/47 (38%), Positives = 28/47 (59%)
Frame = -3
Query: 1355 IKWEQLPTCDNTWELAAVIQDKFPLFPLEDKVKLLGGVLIGLGLSQP 1401
I+W+ LPT +++WE + D FP LED++ L GG + S+P
Sbjct: 477 IRWKDLPTFEDSWEDFCKLLDPFPNHQLEDQLNLQGGRDVANPSSRP 337
>TC18698
Length = 808
Score = 38.5 bits (88), Expect = 0.008
Identities = 21/70 (30%), Positives = 38/70 (54%)
Frame = -2
Query: 595 EKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDLLT 654
+KT + + +Y + VMP GL N +T+Q LM+++ + K V+ +D++V S
Sbjct: 804 KKTTLKINRVNYYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKSSQE*F 625
Query: 655 HTTHLQQVLT 664
H L + L+
Sbjct: 624 HRGDLSRDLS 595
>AV777492
Length = 472
Score = 31.6 bits (70), Expect = 0.92
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +1
Query: 1142 NRCLETYLRCFVGPKPKQ 1159
NRCLE Y RC G +PK+
Sbjct: 406 NRCLEVYFRCLTGARPKK 459
>TC9940 weakly similar to UP|Q9LHB1 (Q9LHB1) Transcription factor X1-like
protein, partial (18%)
Length = 577
Score = 31.2 bits (69), Expect = 1.2
Identities = 34/136 (25%), Positives = 56/136 (41%), Gaps = 9/136 (6%)
Frame = +3
Query: 27 REVTEDERMQATMVALEGRALSWFQWWERCNPSPTWEAFKLAVVRRFQPSMIQNPFELLL 86
R E +M+ E RA WE P W FK++++ +I + E L
Sbjct: 21 RPFVEAMKMKYNEADAEDRASELCSLWEEYLKDPDWHPFKISMIEGKHQEIIDDEDEKLR 200
Query: 87 SLKQVGTVDEYVEEFEKYAGALKEIDHDFVRGIFL--------NGLKEEIKAEVRLIQKS 138
LK + E V +E AL+EI+ G ++ G + K V+++ K
Sbjct: 201 GLK--NEMGEGV--YEAVVTALREINEYNPSGRYITSELWNYKEGRRATSKEGVQVLLKQ 368
Query: 139 -LLIEQKNQIVTKKGV 153
L +QK ++ K G+
Sbjct: 369 WKLYKQKTGML*KPGM 416
>AV779679
Length = 440
Score = 30.8 bits (68), Expect = 1.6
Identities = 14/28 (50%), Positives = 19/28 (67%)
Frame = +3
Query: 540 RLCVDYRALNKVTIPNKFPIPIIEELLD 567
+LC DY L+ VTIPNK +P ++E D
Sbjct: 351 QLCDDYMQLDYVTIPNKSLLPHLDEWSD 434
>AV768479
Length = 462
Score = 30.8 bits (68), Expect = 1.6
Identities = 12/37 (32%), Positives = 20/37 (53%)
Frame = +3
Query: 489 VPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRPSISPF 525
+P RP++C Y K+ + + + G I+PS PF
Sbjct: 156 IPTTRPHRCQWYPKHILHHTIMPRVPHGFIQPSDKPF 266
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,836,078
Number of Sequences: 28460
Number of extensions: 380264
Number of successful extensions: 1771
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1745
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1767
length of query: 1430
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1328
effective length of database: 1,994,680
effective search space: 2648935040
effective search space used: 2648935040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0281a.3


