

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.2
(153 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
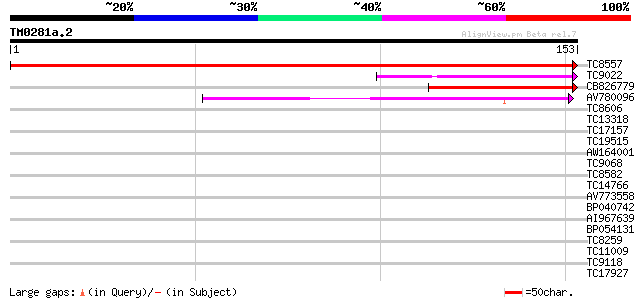
Score E
Sequences producing significant alignments: (bits) Value
TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete 309 1e-85
TC9022 similar to PIR|T46225|T46225 alpha NAC-like protein - Ara... 42 6e-05
CB826779 41 1e-04
AV780096 39 3e-04
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 36 0.003
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 36 0.003
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 36 0.003
TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, pa... 36 0.003
AW164001 34 0.010
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 34 0.010
TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 34 0.013
TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associ... 33 0.017
AV773558 33 0.022
BP040742 33 0.022
AI967639 33 0.028
BP054131 32 0.037
TC8259 similar to UP|HMGL_SOYBN (P26585) HMG1/2-like protein (SB... 32 0.037
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 32 0.049
TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding l... 32 0.049
TC17927 UP|Q8GRY7 (Q8GRY7) bZIP with a Ring-finger motif, complete 32 0.063
>TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete
Length = 1064
Score = 309 bits (792), Expect = 1e-85
Identities = 153/153 (100%), Positives = 153/153 (100%)
Frame = +3
Query: 1 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 60
MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR
Sbjct: 153 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 332
Query: 61 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD 120
DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD
Sbjct: 333 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD 512
Query: 121 EESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
EESDDDDDDEDDTEALLAELEQIKKERAEEKLR
Sbjct: 513 EESDDDDDDEDDTEALLAELEQIKKERAEEKLR 611
>TC9022 similar to PIR|T46225|T46225 alpha NAC-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (75%)
Length = 1083
Score = 41.6 bits (96), Expect = 6e-05
Identities = 21/54 (38%), Positives = 30/54 (54%)
Frame = +3
Query: 100 EDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
+D + V DD D E DE+ DDDD+D+D + L E K+ R+E+K R
Sbjct: 204 DDAPIVEDVKDDDKD-EADDDEDEDDDDEDDDKEDGALGGSEGSKQSRSEKKSR 362
>CB826779
Length = 469
Score = 40.8 bits (94), Expect = 1e-04
Identities = 18/40 (45%), Positives = 26/40 (65%)
Frame = +1
Query: 114 DVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
DV ++DE+ DDD+DDEDD + E K+ R+E+K R
Sbjct: 208 DVHDEADEDEDDDEDDEDDEDGAQGTTEGSKQSRSEKKSR 327
>AV780096
Length = 564
Score = 39.3 bits (90), Expect = 3e-04
Identities = 31/112 (27%), Positives = 47/112 (41%), Gaps = 12/112 (10%)
Frame = +1
Query: 53 ELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADD 112
++ RRN D EE E + + +D D E VARS +DD
Sbjct: 25 DMARRNGSDSEEEEEEQQEEEVHDQIEED----------------DEELEAVARSASSDD 156
Query: 113 SDVEVKSDEESDDDDDDEDD------------TEALLAELEQIKKERAEEKL 152
E ++ DDD++DD +A L EL+Q+KK++ +E L
Sbjct: 157 EPDESGGEDAVAADDDNQDDEGNAADPEISKREKARLKELQQLKKQKIQEIL 312
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 36.2 bits (82), Expect = 0.003
Identities = 16/43 (37%), Positives = 27/43 (62%)
Frame = -2
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
D +D D E + D++ DD+DDDE+D E E +++E E++
Sbjct: 428 DEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDE 300
Score = 32.0 bits (71), Expect = 0.049
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = -2
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTE 134
+ +D + E D+E DDDDD++DD E
Sbjct: 437 NGEDEEDEDGEDQEDDDDDDEDDDEE 360
Score = 30.0 bits (66), Expect = 0.18
Identities = 23/81 (28%), Positives = 36/81 (44%), Gaps = 1/81 (1%)
Frame = -2
Query: 72 SSKNKSYSDDR-DHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDE 130
+S NKS S + G + E D ED D +D + + DE+ D++DD
Sbjct: 509 NSNNKSNSKKAPEGGPGAGAGPEENGEDEEDE------DGEDQEDDDDDDEDDDEEDDGG 348
Query: 131 DDTEALLAELEQIKKERAEEK 151
+D E E E + E +E+
Sbjct: 347 EDEEEEGVEEEDNEDEEEDEE 285
Score = 28.9 bits (63), Expect = 0.41
Identities = 10/26 (38%), Positives = 20/26 (76%)
Frame = -2
Query: 111 DDSDVEVKSDEESDDDDDDEDDTEAL 136
+D + E +E+++D+++DE+D EAL
Sbjct: 347 EDEEEEGVEEEDNEDEEEDEEDEEAL 270
Score = 24.6 bits (52), Expect = 7.7
Identities = 8/33 (24%), Positives = 20/33 (60%)
Frame = -2
Query: 110 ADDSDVEVKSDEESDDDDDDEDDTEALLAELEQ 142
++D + + ++ +DD+ D++DD + E E+
Sbjct: 644 SEDKQHDAEGEDGNDDEGDEDDDGDGPFGEGEE 546
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 36.2 bits (82), Expect = 0.003
Identities = 16/44 (36%), Positives = 26/44 (58%)
Frame = +3
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKL 152
+ DD + E DE+ DDDDDD+DD E + + ++E E++
Sbjct: 258 EEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEV 389
Score = 34.3 bits (77), Expect = 0.010
Identities = 15/33 (45%), Positives = 21/33 (63%)
Frame = +3
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEALLAELE 141
D DD D + +EE DDDDDDE++ + E+E
Sbjct: 300 DDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVE 398
Score = 33.9 bits (76), Expect = 0.013
Identities = 12/26 (46%), Positives = 19/26 (72%)
Frame = +3
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTE 134
+ ++ D E + DE+ DDDDDD+DD +
Sbjct: 252 EEEEDDDEEEDDEDEDDDDDDDDDDD 329
Score = 31.6 bits (70), Expect = 0.063
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = +3
Query: 111 DDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAE 149
DD D + D E ++DDDD+D+ E E+E K+ A+
Sbjct: 300 DDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAK 416
Score = 31.6 bits (70), Expect = 0.063
Identities = 13/45 (28%), Positives = 25/45 (54%)
Frame = +3
Query: 90 LFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
++ +G + V+ D ++ D E + D++ DDDDDD ++ E
Sbjct: 210 IYPDGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEE 344
Score = 28.9 bits (63), Expect = 0.41
Identities = 17/43 (39%), Positives = 23/43 (52%), Gaps = 3/43 (6%)
Frame = +3
Query: 112 DSDVEVKSDEESDD---DDDDEDDTEALLAELEQIKKERAEEK 151
D++ E DEE DD DDDD+DD + E E + EE+
Sbjct: 246 DNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEE 374
Score = 28.9 bits (63), Expect = 0.41
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Frame = +3
Query: 106 RSVDADDSDVEVKSDEESDDD-DDDEDDTEALLAELEQIKKERAEEK 151
+ V D+ + E +EE D+D DDD+DD + E E+ + EE+
Sbjct: 231 KPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEE 371
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 35.8 bits (81), Expect = 0.003
Identities = 19/50 (38%), Positives = 26/50 (52%)
Frame = +2
Query: 85 GKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
G++ E + D ED D D+ + + +EE DDDDDDEDD E
Sbjct: 740 GEAEQSDFEDIEEDDEDG------DEDEDEDDDDDEEEEDDDDDDEDDEE 871
Score = 31.6 bits (70), Expect = 0.063
Identities = 12/37 (32%), Positives = 23/37 (61%)
Frame = +2
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D+E+ D D+ D + + +E+ DDDD+D+++ E
Sbjct: 767 DIEEDDEDGDEDEDEDDDDDEEEEDDDDDDEDDEEGE 877
>TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, partial (3%)
Length = 504
Score = 35.8 bits (81), Expect = 0.003
Identities = 17/29 (58%), Positives = 19/29 (64%)
Frame = +3
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEALL 137
D+D D E DEE DDDDDD DD + LL
Sbjct: 63 DSDPDDAE-SEDEEDDDDDDDGDDEDPLL 146
Score = 32.7 bits (73), Expect = 0.028
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +3
Query: 106 RSVDADDSDVEVKSDEESDDDDDDEDDTE 134
R D D + +S++E DDDDDD+ D E
Sbjct: 48 RPTDDDSDPDDAESEDEEDDDDDDDGDDE 134
>AW164001
Length = 447
Score = 34.3 bits (77), Expect = 0.010
Identities = 20/59 (33%), Positives = 30/59 (49%)
Frame = +1
Query: 94 GTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKL 152
GT V D + ++ D + +V D + DDDDDD+DD + A L + K + E L
Sbjct: 7 GTSDVVGDSVNKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDDEPATLGFVDKPKDEWSL 183
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 34.3 bits (77), Expect = 0.010
Identities = 13/24 (54%), Positives = 17/24 (70%)
Frame = +2
Query: 109 DADDSDVEVKSDEESDDDDDDEDD 132
D DD D + D++ DDDDDD+DD
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDD 154
Score = 33.1 bits (74), Expect = 0.022
Identities = 12/24 (50%), Positives = 17/24 (70%)
Frame = +2
Query: 111 DDSDVEVKSDEESDDDDDDEDDTE 134
DD D + D++ DDDDDD+DD +
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDD 154
Score = 31.6 bits (70), Expect = 0.063
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +2
Query: 109 DADDSDVEVKSDEESDDDDDDEDD 132
D DD D + D++ DDDDDD+ D
Sbjct: 89 DDDDDDDDDDDDDDDDDDDDDDGD 160
Score = 29.6 bits (65), Expect = 0.24
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = +2
Query: 118 KSDEESDDDDDDEDDTE 134
K D++ DDDDDD+DD +
Sbjct: 77 KPDDDDDDDDDDDDDDD 127
Score = 25.4 bits (54), Expect = 4.5
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = +2
Query: 119 SDEESDDDDDDEDDTE 134
S + DDDDDD+DD +
Sbjct: 71 SPKPDDDDDDDDDDDD 118
>TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (65%)
Length = 1341
Score = 33.9 bits (76), Expect = 0.013
Identities = 26/106 (24%), Positives = 46/106 (42%), Gaps = 17/106 (16%)
Frame = +1
Query: 61 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKS- 119
DE EE + K ++ ++D K++ + IV D +++D++V S
Sbjct: 394 DEEEEELMEQENGKPETKTEDAKVAKTAKPAAAAGPSAKQVKIVDPKKDEEEADLDVSSP 573
Query: 120 ----------------DEESDDDDDDEDDTEALLAELEQIKKERAE 149
D+ESDD+ DDE++T +++Q KK E
Sbjct: 574 DVSGYEDDLISADEDSDDESDDESDDEEETPTPAKKVDQGKKRPNE 711
>TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associated
complex alpha chain, partial (58%)
Length = 600
Score = 33.5 bits (75), Expect = 0.017
Identities = 18/55 (32%), Positives = 31/55 (55%), Gaps = 3/55 (5%)
Frame = +2
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELE---QIKKERAEEKLR 153
H+ + + D+ VE +++ D+DDDDEDD E + + K+ R+E+K R
Sbjct: 95 HLEQQKIHDDEPVVEDDDEDDDDEDDDDEDDDNIEGQEGDASGRSKQTRSEKKSR 259
Score = 26.6 bits (57), Expect = 2.0
Identities = 12/40 (30%), Positives = 21/40 (52%)
Frame = +2
Query: 95 TKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
T + E+ + A D E +++ +DDDD++DD E
Sbjct: 62 TAQSQEELLAAHLEQQKIHDDEPVVEDDDEDDDDEDDDDE 181
>AV773558
Length = 469
Score = 33.1 bits (74), Expect = 0.022
Identities = 13/26 (50%), Positives = 19/26 (73%)
Frame = -2
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTE 134
+ DD V V S+++ DDDDDD+DD +
Sbjct: 399 ELDDDFVLVYSEDDDDDDDDDDDDDD 322
Score = 32.0 bits (71), Expect = 0.049
Identities = 14/41 (34%), Positives = 24/41 (58%)
Frame = -2
Query: 91 FLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDED 131
F EG + +D ++ S D DD D + D++ DD+D+E+
Sbjct: 417 FNEGDEELDDDFVLVYSEDDDDDDDDDDDDDDYTDDEDEEE 295
Score = 28.9 bits (63), Expect = 0.41
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = -2
Query: 95 TKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
T D D + D + D V E DDDDDD+DD +
Sbjct: 447 TTEDSPDGLFFNEGDEELDDDFVLVYSEDDDDDDDDDDDD 328
>BP040742
Length = 465
Score = 33.1 bits (74), Expect = 0.022
Identities = 15/28 (53%), Positives = 18/28 (63%)
Frame = -1
Query: 116 EVKSDEESDDDDDDEDDTEALLAELEQI 143
E DE+ +DDDDDEDD L AEL +
Sbjct: 465 EEDDDEDDEDDDDDEDDD*DLCAELNMM 382
>AI967639
Length = 335
Score = 32.7 bits (73), Expect = 0.028
Identities = 22/57 (38%), Positives = 25/57 (43%)
Frame = +1
Query: 76 KSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDD 132
K RD SS EG + D E+ D D DEE DDDDDD+DD
Sbjct: 58 KKRKQRRDISSSS----EGDEEDEEE---------DQGDEYNLEDEEEDDDDDDDDD 189
Score = 30.8 bits (68), Expect = 0.11
Identities = 14/57 (24%), Positives = 29/57 (50%)
Frame = +1
Query: 96 KRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKL 152
+RD+ D ++ + + E+ ++DDDD+DD + ++E K R ++L
Sbjct: 73 RRDISSSSEGDEEDEEEDQGDEYNLEDEEEDDDDDDDDDLSISEDSDSDKPRKVKQL 243
>BP054131
Length = 500
Score = 32.3 bits (72), Expect = 0.037
Identities = 29/88 (32%), Positives = 39/88 (43%), Gaps = 10/88 (11%)
Frame = -2
Query: 55 KRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSD 114
KRR + LEE R+ S +K D G + + G K D E +D D+ +
Sbjct: 415 KRREEKQRLEEGRRKE--SMDKLRMDMEKSGGGTVVLNTGLK-DQEKAKSKSKMDEDEDE 245
Query: 115 VEVKSDEESDDD----------DDDEDD 132
VE DEE DDD D+DED+
Sbjct: 244 VE--EDEEMDDDLVYTEDEDAIDEDEDE 167
>TC8259 similar to UP|HMGL_SOYBN (P26585) HMG1/2-like protein (SB11
protein), partial (92%)
Length = 942
Score = 32.3 bits (72), Expect = 0.037
Identities = 18/72 (25%), Positives = 33/72 (45%)
Frame = +2
Query: 63 LEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEE 122
+ E E+ + +K + D + ++ K+ E A +++ S EV +E
Sbjct: 431 MSEAEKAPYVAKAEKRKADYEKTMKAY-----NKKQAEGPAAADEEESEKSVSEVNDQDE 595
Query: 123 SDDDDDDEDDTE 134
DDD D++DD E
Sbjct: 596 DDDDSDEDDDDE 631
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 32.0 bits (71), Expect = 0.049
Identities = 21/65 (32%), Positives = 29/65 (44%), Gaps = 10/65 (15%)
Frame = +2
Query: 80 DDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKS----------DEESDDDDDD 129
DD D G+ +G + D +D D D+ D E + D + DDDDDD
Sbjct: 32 DDDDDGEDD----DGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDD 199
Query: 130 EDDTE 134
+DD E
Sbjct: 200 DDDEE 214
Score = 26.9 bits (58), Expect = 1.6
Identities = 16/50 (32%), Positives = 24/50 (48%)
Frame = +2
Query: 80 DDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDD 129
DD + + L GT+ V + A + SD E + DE D+D+DD
Sbjct: 203 DDEEEEEEEDL---GTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDD 343
Score = 26.2 bits (56), Expect = 2.7
Identities = 13/26 (50%), Positives = 15/26 (57%)
Frame = +2
Query: 107 SVDADDSDVEVKSDEESDDDDDDEDD 132
S D DD D D E DD D+D+DD
Sbjct: 20 SGDEDDDD-----DGEDDDGDEDDDD 82
Score = 25.0 bits (53), Expect = 5.9
Identities = 9/23 (39%), Positives = 14/23 (60%)
Frame = +2
Query: 112 DSDVEVKSDEESDDDDDDEDDTE 134
+S + DE+ DDD +D+D E
Sbjct: 2 NSRTRMSGDEDDDDDGEDDDGDE 70
Score = 25.0 bits (53), Expect = 5.9
Identities = 8/23 (34%), Positives = 15/23 (64%)
Frame = +2
Query: 108 VDADDSDVEVKSDEESDDDDDDE 130
+ D+ D + D++ D+DDDD+
Sbjct: 17 MSGDEDDDDDGEDDDGDEDDDDD 85
>TC9118 similar to UP|Q84JE4 (Q84JE4) Squamosa promoter binding
like-protein, partial (16%)
Length = 779
Score = 32.0 bits (71), Expect = 0.049
Identities = 19/83 (22%), Positives = 37/83 (43%)
Frame = +2
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
++ ++E KR ++RD R ++H S ++ S DRD H + +D + A+
Sbjct: 35 EEEEEEEKREDVRDHKRSR-KKHRSHRSSHTSRDRDRDGRKHKRRHSSSKDKYSYRGAKH 211
Query: 108 VDADDSDVEVKSDEESDDDDDDE 130
+DD + D D++
Sbjct: 212 GISDDEHQHSSHHHKYDSSSDEK 280
>TC17927 UP|Q8GRY7 (Q8GRY7) bZIP with a Ring-finger motif, complete
Length = 1793
Score = 31.6 bits (70), Expect = 0.063
Identities = 16/42 (38%), Positives = 24/42 (57%)
Frame = +1
Query: 112 DSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
+S E E DDDDDDEDD + L E++ + R + +L+
Sbjct: 718 ESHKESSQMEGDDDDDDDEDDVDDLENEVKYGQGNRMKARLQ 843
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.306 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,795,479
Number of Sequences: 28460
Number of extensions: 20087
Number of successful extensions: 866
Number of sequences better than 10.0: 149
Number of HSP's better than 10.0 without gapping: 334
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 559
length of query: 153
length of database: 4,897,600
effective HSP length: 82
effective length of query: 71
effective length of database: 2,563,880
effective search space: 182035480
effective search space used: 182035480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0281a.2


