

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0276.6
(483 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
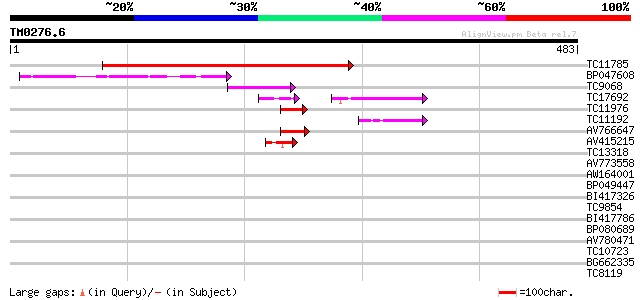
Score E
Sequences producing significant alignments: (bits) Value
TC11785 homologue to UP|Q9ZVC2 (Q9ZVC2) At2g41370 protein, parti... 433 e-122
BP047608 45 3e-05
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 45 3e-05
TC17692 weakly similar to UP|Q8LLW2 (Q8LLW2) Receptor-like kinas... 35 3e-05
TC11976 similar to GB|AAP12856.1|30017245|BT006207 At3g02340 {Ar... 41 4e-04
TC11192 similar to GB|AAO63323.1|28950799|BT005259 At2g03430 {Ar... 41 4e-04
AV766647 40 8e-04
AV415215 40 8e-04
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 39 0.002
AV773558 39 0.002
AW164001 38 0.003
BP049447 34 0.003
BI417326 38 0.004
TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1 (AT3g1... 38 0.004
BI417786 38 0.004
BP080689 38 0.004
AV780471 37 0.005
TC10723 similar to GB|AAB61117.1|2194142|F20P5 ESTs gb|N38288,gb... 37 0.005
BG662335 37 0.007
TC8119 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protei... 37 0.007
>TC11785 homologue to UP|Q9ZVC2 (Q9ZVC2) At2g41370 protein, partial (42%)
Length = 644
Score = 433 bits (1113), Expect = e-122
Identities = 214/214 (100%), Positives = 214/214 (100%)
Frame = +1
Query: 80 GGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTL 139
GGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTL
Sbjct: 1 GGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTL 180
Query: 140 AAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKSGLP 199
AAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKSGLP
Sbjct: 181 AAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKSGLP 360
Query: 200 PEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRM 259
PEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRM
Sbjct: 361 PEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRM 540
Query: 260 RRALDSSDVELVKLMVMGEGLNLDEALALPYAVE 293
RRALDSSDVELVKLMVMGEGLNLDEALALPYAVE
Sbjct: 541 RRALDSSDVELVKLMVMGEGLNLDEALALPYAVE 642
>BP047608
Length = 565
Score = 45.1 bits (105), Expect = 3e-05
Identities = 44/181 (24%), Positives = 75/181 (41%)
Frame = +2
Query: 9 SLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDS 68
++ LD+ L + D+ F V+ AH+ +LAARS FR F GP
Sbjct: 110 AMGLDFKELF-ESEVGCDIVFKVKSESFKAHKVVLAARSPVFRAQFFGP----------- 253
Query: 69 PGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHC 128
V P+ + V + +F ML F+YS ++ + + P C A H
Sbjct: 254 -----VGDPKQEEAV-VEDIEPFIFKAMLLFIYSDKLPDIYEVMGSVPMC-SYAVMMQHL 412
Query: 129 TSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTT 188
+A DL + +++L LL + +L ++ +I++V L + + QL
Sbjct: 413 VAAADL----------YNIDRLKLLCESKLC---DEITIDNVATSLALAEQHHCPQLKAI 553
Query: 189 C 189
C
Sbjct: 554 C 556
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 44.7 bits (104), Expect = 3e-05
Identities = 21/58 (36%), Positives = 26/58 (44%)
Frame = -2
Query: 186 WTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHH 243
W C + + AKH P + +K + R RR HHHHHHHHH HH
Sbjct: 278 WGPCRYSQRQQNPRQNQGAKHRPKEYPSKRCQCRSLVVSCRRRHHHHHHHHHHHHHHH 105
>TC17692 weakly similar to UP|Q8LLW2 (Q8LLW2) Receptor-like kinase
Xa21-binding protein 3, partial (19%)
Length = 482
Score = 35.4 bits (80), Expect = 0.021
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = +2
Query: 232 HHHHHHHHHGHHDMGAAADLEDQ 254
H HHHHHH HH G+ + L+ Q
Sbjct: 137 HLSHHHHHHHHHHNGSGSQLQRQ 205
Score = 33.9 bits (76), Expect(2) = 3e-05
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +2
Query: 213 AKIEELRLKSTLARRSLIPHHHHHHHHHGHHDMGA 247
AK E++ L + HHHHHHHHH H+ G+
Sbjct: 101 AKSREVKFNKPLH----LSHHHHHHHHH-HNGSGS 190
Score = 30.0 bits (66), Expect(2) = 3e-05
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 4/86 (4%)
Frame = +3
Query: 275 VMGEGL----NLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMV 330
+MG+GL N D L AV+ ++V ALLE + + + LHIAA
Sbjct: 174 IMGQGLSCRGNHDHGLFT--AVQQGDLQIVTALLEQDPTLFHQTTLYDRHSALHIAAANG 347
Query: 331 SPDMVAVLLDHHADPNVRTVDGVTPL 356
++++ LLD +P++ TPL
Sbjct: 348 QIEILSRLLDGSVNPDILNRQKQTPL 425
>TC11976 similar to GB|AAP12856.1|30017245|BT006207 At3g02340 {Arabidopsis
thaliana;}, partial (11%)
Length = 530
Score = 41.2 bits (95), Expect = 4e-04
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = -3
Query: 231 PHHHHHHHHHGHHDMGAAADLED 253
PHHHHHHHHH HH + + +
Sbjct: 225 PHHHHHHHHHHHHHFTGSVQISE 157
Score = 33.1 bits (74), Expect = 0.10
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = -3
Query: 216 EELRLKSTLARRSLIPHHHHHHHHHGHH 243
+ L L + LA HHHHHHH HH
Sbjct: 276 KNLNLTA*LAGAEKFQKPHHHHHHHHHH 193
>TC11192 similar to GB|AAO63323.1|28950799|BT005259 At2g03430 {Arabidopsis
thaliana;}, partial (70%)
Length = 597
Score = 41.2 bits (95), Expect = 4e-04
Identities = 25/59 (42%), Positives = 34/59 (57%)
Frame = +3
Query: 298 EVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPL 356
E+V+ALL GA DVN +G G+T LH AA + +L+ H A N++ G TPL
Sbjct: 336 EIVEALLSKGA-DVNLKNG-GGRTALHYAASKGRVKIAEILISHDAKINIKDKVGCTPL 506
>AV766647
Length = 349
Score = 40.0 bits (92), Expect = 8e-04
Identities = 14/25 (56%), Positives = 15/25 (60%)
Frame = -3
Query: 231 PHHHHHHHHHGHHDMGAAADLEDQK 255
PH HHHHHHH HH+ G D K
Sbjct: 332 PHPHHHHHHHHHHEHGGGCCSGDAK 258
Score = 30.0 bits (66), Expect = 0.88
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
H H HHHHH HH
Sbjct: 335 HPHPHHHHHHHH 300
Score = 26.9 bits (58), Expect = 7.4
Identities = 8/12 (66%), Positives = 8/12 (66%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
H H H HHH HH
Sbjct: 341 HQHPHPHHHHHH 306
>AV415215
Length = 430
Score = 40.0 bits (92), Expect = 8e-04
Identities = 17/31 (54%), Positives = 22/31 (70%), Gaps = 4/31 (12%)
Frame = +3
Query: 219 RLKSTLARRSLIP----HHHHHHHHHGHHDM 245
RLK+ L ++L+P HHHHHHHHH HH +
Sbjct: 123 RLKTPL--QNLLPPFQTHHHHHHHHHHHHHL 209
Score = 32.7 bits (73), Expect = 0.14
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHH HH HH
Sbjct: 189 HHHHHHLHHIHH 224
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 38.9 bits (89), Expect = 0.002
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = -1
Query: 229 LIPHHHHHHHHHGHH 243
L+P HHHHHHHH HH
Sbjct: 342 LLPPHHHHHHHHHHH 298
Score = 36.2 bits (82), Expect = 0.012
Identities = 15/35 (42%), Positives = 18/35 (50%), Gaps = 9/35 (25%)
Frame = -1
Query: 220 LKSTLARRSLIPHHHHHH---------HHHGHHDM 245
L+ L L+PHHHHHH HHH HH +
Sbjct: 399 LRLLLLHFPLLPHHHHHHLLLPPHHHHHHHHHHHL 295
Score = 33.9 bits (76), Expect = 0.061
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = -1
Query: 232 HHHHHHHHHGH 242
HHHHHHHHH H
Sbjct: 324 HHHHHHHHHLH 292
Score = 33.5 bits (75), Expect = 0.079
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = -1
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH H
Sbjct: 327 HHHHHHHHHHLH 292
Score = 32.7 bits (73), Expect = 0.14
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = -1
Query: 232 HHHHHHHHHGHH 243
HHHHHHH H HH
Sbjct: 318 HHHHHHHLHLHH 283
>AV773558
Length = 469
Score = 38.5 bits (88), Expect = 0.002
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = +3
Query: 229 LIPHHHHHHHHHGHH 243
L HHHHHHHHH HH
Sbjct: 315 LCSHHHHHHHHHHHH 359
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 327 HHHHHHHHHHHH 362
Score = 35.4 bits (80), Expect = 0.021
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH H+
Sbjct: 336 HHHHHHHHHPHY 371
Score = 33.9 bits (76), Expect = 0.061
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH H
Sbjct: 333 HHHHHHHHHHPH 368
Score = 29.6 bits (65), Expect = 1.1
Identities = 11/18 (61%), Positives = 12/18 (66%), Gaps = 3/18 (16%)
Frame = +3
Query: 229 LIPHHHH---HHHHHGHH 243
L+ H HH HHHHH HH
Sbjct: 294 LLLHPHHLCSHHHHHHHH 347
>AW164001
Length = 447
Score = 38.1 bits (87), Expect = 0.003
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = -3
Query: 230 IPHHHHHHHHHGHH 243
+ HHHHHHHHH HH
Sbjct: 136 VRHHHHHHHHHHHH 95
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 127 HHHHHHHHHHHH 92
Score = 35.0 bits (79), Expect = 0.027
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = -3
Query: 232 HHHHHHHHHGHHD 244
HHHHHHHHH H +
Sbjct: 124 HHHHHHHHHHHQN 86
Score = 31.6 bits (70), Expect = 0.30
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHH H
Sbjct: 115 HHHHHHHHQNPH 80
>BP049447
Length = 478
Score = 34.3 bits (77), Expect(2) = 0.003
Identities = 10/11 (90%), Positives = 11/11 (99%)
Frame = +3
Query: 229 LIPHHHHHHHH 239
L+PHHHHHHHH
Sbjct: 162 LLPHHHHHHHH 194
Score = 30.0 bits (66), Expect(2) = 0.055
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +3
Query: 225 ARRSLIPHHHHHHHHH 240
AR + + HHHHHHHH
Sbjct: 147 ARINQLLPHHHHHHHH 194
Score = 26.9 bits (58), Expect = 7.4
Identities = 9/13 (69%), Positives = 10/13 (76%)
Frame = +1
Query: 228 SLIPHHHHHHHHH 240
SL HHHHHHH+
Sbjct: 214 SLQDLHHHHHHHN 252
Score = 22.7 bits (47), Expect(2) = 0.003
Identities = 6/8 (75%), Positives = 7/8 (87%)
Frame = +1
Query: 237 HHHHGHHD 244
HHHH HH+
Sbjct: 229 HHHHHHHN 252
Score = 22.7 bits (47), Expect(2) = 0.055
Identities = 6/8 (75%), Positives = 7/8 (87%)
Frame = +1
Query: 236 HHHHHGHH 243
HHHHH H+
Sbjct: 229 HHHHHHHN 252
>BI417326
Length = 478
Score = 37.7 bits (86), Expect = 0.004
Identities = 36/109 (33%), Positives = 48/109 (44%)
Frame = +2
Query: 248 AADLEDQKIRRMRRALDSSDVELVKLMVMGEGLNLDEALALPYAVENCSREVVKALLELG 307
A D ED A D+S E V D A+ L Y E + LL+ G
Sbjct: 95 APDHEDSSAVAAAAADDASVTEAV-----------DPAVRLMYLANEGDLEGITELLDAG 241
Query: 308 AADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADPNVRTVDGVTPL 356
+ DVN+ G+T LH+AA D+V +LL AD + + G TPL
Sbjct: 242 S-DVNFRD-IDGRTALHVAACHRRTDVVDLLLQRGADVDTQDRWGSTPL 382
Score = 30.4 bits (67), Expect = 0.67
Identities = 31/105 (29%), Positives = 48/105 (45%), Gaps = 3/105 (2%)
Frame = +2
Query: 244 DMGAAADLEDQKIRRMRRALDSSDVELVKLMVMGEGLN---LDEALALPYAVENCSREVV 300
D + + D +R M A + + +L+ G +N +D AL A + +VV
Sbjct: 140 DDASVTEAVDPAVRLMYLANEGDLEGITELLDAGSDVNFRDIDGRTALHVAACHRRTDVV 319
Query: 301 KALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHADP 345
LL+ GA DV+ G TPL A + D+V +L H A+P
Sbjct: 320 DLLLQRGA-DVD-TQDRWGSTPLADALYYKNHDVVKLLEKHGAEP 448
>TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1
(AT3g17100/K14A17_22_), partial (43%)
Length = 1108
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 11 HHHHHHHHHHHH 46
Score = 36.6 bits (83), Expect = 0.009
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +2
Query: 231 PHHHHHHHHHGHH 243
P HHHHHHHH HH
Sbjct: 5 PTHHHHHHHHHHH 43
>BI417786
Length = 513
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 119 HHHHHHHHHHHH 154
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 113 HHHHHHHHHHHH 148
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 116 HHHHHHHHHHHH 151
Score = 37.0 bits (84), Expect = 0.007
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +2
Query: 231 PHHHHHHHHHGHH 243
P HHHHHHHH HH
Sbjct: 107 PSHHHHHHHHHHH 145
Score = 32.0 bits (71), Expect = 0.23
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = +2
Query: 223 TLARRSLIPHHHHHHHHHGHH 243
TL+ + HHHHHHH HH
Sbjct: 80 TLSHLTSYHPSHHHHHHHHHH 142
>BP080689
Length = 326
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +1
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 4 HHHHHHHHHHHH 39
Score = 37.7 bits (86), Expect = 0.004
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +1
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 1 HHHHHHHHHHHH 36
Score = 37.4 bits (85), Expect = 0.005
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 232 HHHHHHHHHGHHDMG 246
HHHHHHHHH H+ +G
Sbjct: 13 HHHHHHHHHAHNFIG 57
Score = 34.3 bits (77), Expect = 0.046
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +1
Query: 232 HHHHHHHHHGHHD 244
HHHHHHHHH H+
Sbjct: 10 HHHHHHHHHHAHN 48
>AV780471
Length = 527
Score = 37.4 bits (85), Expect = 0.005
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = +1
Query: 214 KIEELRLKSTLARRSLIPHHHHHHHHH 240
K+ T RRS HHHHHHHHH
Sbjct: 145 KVSSSTTGRTHRRRSFSDHHHHHHHHH 225
Score = 33.5 bits (75), Expect = 0.079
Identities = 11/17 (64%), Positives = 11/17 (64%)
Frame = +1
Query: 226 RRSLIPHHHHHHHHHGH 242
RR HHHHHHHH H
Sbjct: 178 RRRSFSDHHHHHHHHHH 228
>TC10723 similar to GB|AAB61117.1|2194142|F20P5 ESTs
gb|N38288,gb|T43486,gb|AA395242 come from this gene.
{Arabidopsis thaliana;}, partial (49%)
Length = 1008
Score = 37.4 bits (85), Expect = 0.005
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 2/28 (7%)
Frame = -2
Query: 232 HHHHHHHHHGHHDMGAA--ADLEDQKIR 257
+HHHHHHHH HH A ++E++K++
Sbjct: 689 NHHHHHHHHHHHSKS*AYPTNMEEEKLQ 606
>BG662335
Length = 336
Score = 37.0 bits (84), Expect = 0.007
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = +1
Query: 219 RLKSTLARRSLIPHHHHHHHHHGHH 243
+LK + PH HHHHHHH HH
Sbjct: 262 QLKGKQLHQHQHPHPHHHHHHHHHH 336
Score = 29.3 bits (64), Expect = 1.5
Identities = 10/24 (41%), Positives = 13/24 (53%)
Frame = +1
Query: 220 LKSTLARRSLIPHHHHHHHHHGHH 243
++ L + L H H H HHH HH
Sbjct: 253 MRKQLKGKQLHQHQHPHPHHHHHH 324
>TC8119 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protein-like,
partial (85%)
Length = 1249
Score = 37.0 bits (84), Expect = 0.007
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = +1
Query: 232 HHHHHHHHHGHHDM 245
H+HHHHHHH HH M
Sbjct: 124 HYHHHHHHHHHHHM 165
Score = 34.3 bits (77), Expect = 0.046
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +1
Query: 228 SLIPHHHHHHHHHGHH 243
SLI H+HHHHHH HH
Sbjct: 109 SLIFLHYHHHHHHHHH 156
Score = 34.3 bits (77), Expect = 0.046
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +1
Query: 232 HHHHHHHHHGH 242
HHHHHHHHH H
Sbjct: 136 HHHHHHHHHMH 168
Score = 33.9 bits (76), Expect = 0.061
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +1
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH H
Sbjct: 133 HHHHHHHHHHMH 168
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,114,162
Number of Sequences: 28460
Number of extensions: 183142
Number of successful extensions: 7164
Number of sequences better than 10.0: 327
Number of HSP's better than 10.0 without gapping: 2654
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4883
length of query: 483
length of database: 4,897,600
effective HSP length: 94
effective length of query: 389
effective length of database: 2,222,360
effective search space: 864498040
effective search space used: 864498040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0276.6


