

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
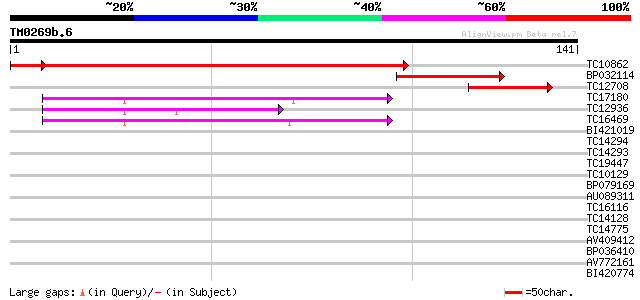
Query= TM0269b.6
(141 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10862 UP|RK2_LOTJA (Q9B1H9) Chloroplast 50S ribosomal protein ... 198 9e-54
BP032114 44 8e-06
TC12708 NADH dehydrogenase ND2 [Lotus japonicus] 44 1e-05
TC17180 homologue to UP|Q9FEZ7 (Q9FEZ7) Ribosomal protein L2, pa... 42 4e-05
TC12936 homologue to UP|Q9FEZ7 (Q9FEZ7) Ribosomal protein L2, pa... 40 1e-04
TC16469 homologue to UP|Q9FEZ7 (Q9FEZ7) Ribosomal protein L2, co... 39 3e-04
BI421019 30 0.21
TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 29 0.35
TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 29 0.35
TC19447 weakly similar to UP|Q8LPN7 (Q8LPN7) AT3g19950/MPN9_19, ... 28 0.60
TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%) 28 0.78
BP079169 27 1.0
AU089311 27 1.0
TC16116 weakly similar to UP|O80798 (O80798) T8F5.4 protein (Exp... 27 1.3
TC14128 similar to AAS09980 (AAS09980) MYB transcription factor,... 27 1.3
TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest c... 27 1.3
AV409412 27 1.7
BP036410 26 2.3
AV772161 26 2.3
BI420774 26 3.0
>TC10862 UP|RK2_LOTJA (Q9B1H9) Chloroplast 50S ribosomal protein L2,
complete
Length = 837
Score = 198 bits (504), Expect(2) = 9e-54
Identities = 90/91 (98%), Positives = 91/91 (99%)
Frame = -2
Query: 9 PPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSPEGN 68
PPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSPEGN
Sbjct: 710 PPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSPEGN 531
Query: 69 FNVADFPSFAISFATAPAALANFPPFPNVIT 99
FNVADFPSFAISFATAPAALANFPPFPNVI+
Sbjct: 530 FNVADFPSFAISFATAPAALANFPPFPNVIS 438
Score = 26.6 bits (57), Expect(2) = 9e-54
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = -1
Query: 1 NGGPPFTPP 9
NGGPPFTPP
Sbjct: 729 NGGPPFTPP 703
>BP032114
Length = 509
Score = 44.3 bits (103), Expect = 8e-06
Identities = 19/27 (70%), Positives = 21/27 (77%)
Frame = +3
Query: 97 VITHFFINRTFYKRHKTKRDNPSRAKL 123
+ THFFINRTFYKRHKTKR N + L
Sbjct: 366 IYTHFFINRTFYKRHKTKRANNKQNSL 446
>TC12708 NADH dehydrogenase ND2 [Lotus japonicus]
Length = 1164
Score = 43.9 bits (102), Expect = 1e-05
Identities = 20/21 (95%), Positives = 21/21 (99%)
Frame = +3
Query: 115 RDNPSRAKL*KVPFKIKQFHR 135
R+NPSRAKL*KVPFKIKQFHR
Sbjct: 1011 RNNPSRAKL*KVPFKIKQFHR 1073
>TC17180 homologue to UP|Q9FEZ7 (Q9FEZ7) Ribosomal protein L2, partial (54%)
Length = 644
Score = 42.0 bits (97), Expect = 4e-05
Identities = 35/94 (37%), Positives = 45/94 (47%), Gaps = 7/94 (7%)
Frame = -1
Query: 9 PPPWGWSTGFITTPLTTGR-----LPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRT 63
PPP G STGFI TP T G+ L PA + F P TCP +A Q L
Sbjct: 287 PPP*GCSTGFIATPRTFGQQFLFTLNLW*ALPAFRRGFSVLPPPATCPIIALQSLGTIFF 108
Query: 64 SPEGNF--NVADFPSFAISFATAPAALANFPPFP 95
+P+G+F V+ ++ A +P LAN P P
Sbjct: 107 APDGSFILEVSLSGL*LMTMA*SPEHLANTPRSP 6
>TC12936 homologue to UP|Q9FEZ7 (Q9FEZ7) Ribosomal protein L2, partial (48%)
Length = 491
Score = 40.4 bits (93), Expect = 1e-04
Identities = 29/65 (44%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Frame = -1
Query: 9 PPPWGWSTGFITTPLTTGR--LPSQHFDPALPKL---FWFTPTFPTCPTVAEQFLDIKRT 63
PPP G STGFI TP T G+ L + + ALP L F P TCP +A Q L
Sbjct: 239 PPP*GCSTGFIATPRTLGQQFLFTLNLW*ALPALRSGFSVLPPPATCPIIALQSLGTIFF 60
Query: 64 SPEGN 68
+PEG+
Sbjct: 59 APEGS 45
>TC16469 homologue to UP|Q9FEZ7 (Q9FEZ7) Ribosomal protein L2, complete
Length = 1003
Score = 38.9 bits (89), Expect = 3e-04
Identities = 34/94 (36%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Frame = -1
Query: 9 PPPWGWSTGFITTPLTTGR-----LPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRT 63
PPP G STGFI TP T G+ L PAL + F P TCP +A L
Sbjct: 685 PPP*GCSTGFIATPRTLGQQFLFTLNLW*ALPALRRGFSVLPPPATCPIMALHPLGTIFF 506
Query: 64 SPEGN--FNVADFPSFAISFATAPAALANFPPFP 95
+P+G+ V+ + I+ A +P LA P P
Sbjct: 505 APDGSLILEVSLSGLWLITIA*SPEHLAKAPRSP 404
>BI421019
Length = 486
Score = 29.6 bits (65), Expect = 0.21
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 1/44 (2%)
Frame = -2
Query: 9 PPPWGWSTGFITTPLTTGRLPSQHFDPAL-PKLFWFTPTFPTCP 51
PPP S +T PL GR+P+ A+ +L+W T F + P
Sbjct: 149 PPPGSPSHPSLTAPLPVGRVPAPPSAVAMGRRLWWLTMPFSSSP 18
>TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (65%)
Length = 1192
Score = 28.9 bits (63), Expect = 0.35
Identities = 9/15 (60%), Positives = 10/15 (66%)
Frame = -3
Query: 2 GGPPFTPPPPWGWST 16
GG P PPPPW W +
Sbjct: 401 GG*PLPPPPPWEWDS 357
>TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (18%)
Length = 540
Score = 28.9 bits (63), Expect = 0.35
Identities = 9/15 (60%), Positives = 10/15 (66%)
Frame = -1
Query: 2 GGPPFTPPPPWGWST 16
GG P PPPPW W +
Sbjct: 414 GG*PLPPPPPWEWDS 370
>TC19447 weakly similar to UP|Q8LPN7 (Q8LPN7) AT3g19950/MPN9_19, partial
(22%)
Length = 534
Score = 28.1 bits (61), Expect = 0.60
Identities = 27/92 (29%), Positives = 40/92 (43%), Gaps = 6/92 (6%)
Frame = -3
Query: 5 PFTPPPPWGWSTGFITTPLTTGRLPSQHFD-PALPKLFWFTPTFPTCP-----TVAEQFL 58
P PPPP + + PL + S + PALP+L TPT + P T ++ +L
Sbjct: 475 PPPPPPPHPPLSPSPSPPLLNRSMISPPYSAPALPRLSALTPTTRSTPLRSSRTTSKPWL 296
Query: 59 DIKRTSPEGNFNVADFPSFAISFATAPAALAN 90
TS + A P + S AT A+ +
Sbjct: 295 PEAPTSSS*SRTPATLPERSGSLAT*TMAITS 200
>TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%)
Length = 562
Score = 27.7 bits (60), Expect = 0.78
Identities = 17/52 (32%), Positives = 20/52 (37%)
Frame = +1
Query: 1 NGGPPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPT 52
N PP+TP PP TP T + P P P P PT P+
Sbjct: 154 NNHPPYTPTPP--------KTPSPTSQPPYIPLPPKTPSPISQPPHVPTPPS 285
Score = 26.6 bits (57), Expect = 1.7
Identities = 18/56 (32%), Positives = 22/56 (39%), Gaps = 3/56 (5%)
Frame = +1
Query: 4 PPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFW---FTPTFPTCPTVAEQ 56
PP+TP PP TP T + P P P +TPT P P+ Q
Sbjct: 307 PPYTPTPP--------KTPSPTSQPPHTPTPPNTPSPISQPPYTPTPPKTPSPTNQ 450
>BP079169
Length = 356
Score = 27.3 bits (59), Expect = 1.0
Identities = 10/32 (31%), Positives = 20/32 (62%)
Frame = -3
Query: 16 TGFITTPLTTGRLPSQHFDPALPKLFWFTPTF 47
T F+++ ++ LP++HF L +F + PT+
Sbjct: 336 TSFLSSSTSSTTLPNRHFAARLLSIFQYQPTY 241
>AU089311
Length = 344
Score = 27.3 bits (59), Expect = 1.0
Identities = 12/27 (44%), Positives = 13/27 (47%)
Frame = -3
Query: 4 PPFTPPPPWGWSTGFITTPLTTGRLPS 30
PPF PPP W I PL + PS
Sbjct: 318 PPFFPPPSISWFGTIILLPLRSPEPPS 238
>TC16116 weakly similar to UP|O80798 (O80798) T8F5.4 protein (Expressed
protein) (At1g65270), partial (34%)
Length = 677
Score = 26.9 bits (58), Expect = 1.3
Identities = 15/38 (39%), Positives = 17/38 (44%)
Frame = +2
Query: 3 GPPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKL 40
G F PPP S+ F TPL T P+ P KL
Sbjct: 260 GRDFPTPPPIPRSSSFSNTPLETPISPTPATSPLASKL 373
>TC14128 similar to AAS09980 (AAS09980) MYB transcription factor, partial
(36%)
Length = 423
Score = 26.9 bits (58), Expect = 1.3
Identities = 17/43 (39%), Positives = 21/43 (48%)
Frame = +2
Query: 15 STGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQF 57
ST +TTPLTT P P P L +P+ PT +E F
Sbjct: 140 STSTLTTPLTTKNTPPATPPPTKPLL--PSPSSSPTPTASEGF 262
>TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest chain
(isoform B1) - soybean (fragment) {Glycine
max;} , partial (22%)
Length = 848
Score = 26.9 bits (58), Expect = 1.3
Identities = 17/62 (27%), Positives = 25/62 (39%)
Frame = +1
Query: 4 PPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRT 63
P ++P P T P + PS + P+ P+L +P PT P + T
Sbjct: 64 PSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPT 243
Query: 64 SP 65
SP
Sbjct: 244 SP 249
>AV409412
Length = 426
Score = 26.6 bits (57), Expect = 1.7
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 5/87 (5%)
Frame = +1
Query: 4 PPFTPPPPWGWSTGFI----TTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLD 59
PP PPPP+ W ++ P ++ PS +PA P+ P+ P L
Sbjct: 79 PPPPPPPPYFWCCHWLF*ASPPPPSSA*TPS*PTNPASTLAPPTIPSSPSLPPSRNPSL- 255
Query: 60 IKRTSPEGNFNV-ADFPSFAISFATAP 85
+ SP + ++ + PS + S AT+P
Sbjct: 256 LHHLSPSPSLSLPSPSPSPSTSSATSP 336
>BP036410
Length = 550
Score = 26.2 bits (56), Expect = 2.3
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = -2
Query: 3 GPPFTPPPPW 12
GPP PPPPW
Sbjct: 180 GPPSPPPPPW 151
>AV772161
Length = 523
Score = 26.2 bits (56), Expect = 2.3
Identities = 13/37 (35%), Positives = 18/37 (48%), Gaps = 2/37 (5%)
Frame = -1
Query: 12 WGWSTG--FITTPLTTGRLPSQHFDPALPKLFWFTPT 46
W W+ G TT L T ++PS+ LP + W T
Sbjct: 511 WLWNCGPDLNTTCLPTMKIPSRRSRSMLPSILWLVIT 401
>BI420774
Length = 423
Score = 25.8 bits (55), Expect = 3.0
Identities = 10/19 (52%), Positives = 12/19 (62%)
Frame = +1
Query: 4 PPFTPPPPWGWSTGFITTP 22
PPF P PP+ T F T+P
Sbjct: 67 PPFPPSPPYSPLTTFSTSP 123
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.329 0.144 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,385,847
Number of Sequences: 28460
Number of extensions: 55072
Number of successful extensions: 654
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 648
length of query: 141
length of database: 4,897,600
effective HSP length: 81
effective length of query: 60
effective length of database: 2,592,340
effective search space: 155540400
effective search space used: 155540400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0269b.6


