

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
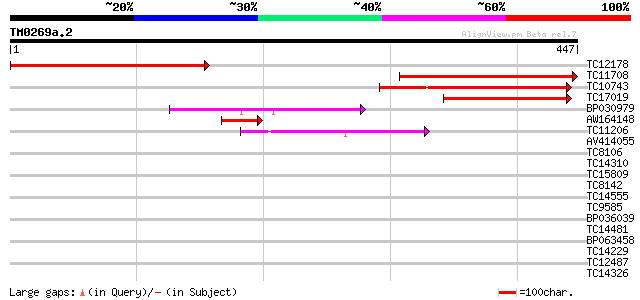
Query= TM0269a.2
(447 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12178 similar to UP|P93557 (P93557) Mitotic cyclin, partial (27%) 305 8e-84
TC11708 similar to UP|O82718 (O82718) Cyclin, partial (31%) 288 2e-78
TC10743 similar to UP|Q8H955 (Q8H955) B1 type cyclin, partial (21%) 147 3e-36
TC17019 similar to UP|Q40223 (Q40223) Cyclin, partial (21%) 111 3e-25
BP030979 85 3e-17
AW164148 55 3e-08
TC11206 similar to UP|Q84V88 (Q84V88) Cyclin D, partial (43%) 44 4e-05
AV414055 32 0.28
TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cu... 31 0.36
TC14310 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, partial (92%) 31 0.36
TC15809 weakly similar to PIR|JN0747|JN0747 histone H1-I - Volvo... 30 0.62
TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete 30 1.1
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 30 1.1
TC9585 similar to UP|O24078 (O24078) Protein phosphatase 2C, par... 29 1.4
BP036039 28 2.3
TC14481 similar to UP|PSBY_SPIOL (P80470) Photosystem II core co... 28 2.3
BP063458 28 4.0
TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%) 28 4.0
TC12487 similar to UP|Q9SAL0 (Q9SAL0) T8K14.20 protein (At1g7938... 28 4.0
TC14326 weakly similar to UP|EXLP_TOBAC (Q03211) Pistil-specific... 28 4.0
>TC12178 similar to UP|P93557 (P93557) Mitotic cyclin, partial (27%)
Length = 554
Score = 305 bits (782), Expect = 8e-84
Identities = 155/157 (98%), Positives = 155/157 (98%)
Frame = +1
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR 60
MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR
Sbjct: 82 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNLDPVRGIEVKPNRPITR 261
Query: 61 SFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKP 120
S CAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKP
Sbjct: 262 SVCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKP 441
Query: 121 VVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTA 157
VVEVIEISPDEQIKKEKSVQKKKEDSKK TLTSVLTA
Sbjct: 442 VVEVIEISPDEQIKKEKSVQKKKEDSKKHTLTSVLTA 552
>TC11708 similar to UP|O82718 (O82718) Cyclin, partial (31%)
Length = 668
Score = 288 bits (736), Expect = 2e-78
Identities = 140/140 (100%), Positives = 140/140 (100%)
Frame = +2
Query: 308 IILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAAS 367
IILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAAS
Sbjct: 2 IILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAAS 181
Query: 368 AVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPE 427
AVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPE
Sbjct: 182 AVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPE 361
Query: 428 RGAVAVLPPAKNLMPQEASS 447
RGAVAVLPPAKNLMPQEASS
Sbjct: 362 RGAVAVLPPAKNLMPQEASS 421
>TC10743 similar to UP|Q8H955 (Q8H955) B1 type cyclin, partial (21%)
Length = 554
Score = 147 bits (372), Expect = 3e-36
Identities = 74/153 (48%), Positives = 107/153 (69%), Gaps = 1/153 (0%)
Frame = +1
Query: 292 LSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGM 351
+S+ AY+ QI ME+ IL +L+W LTVPTP+VFL R + + PD+ + N+A FL+EL +
Sbjct: 1 ISNDAYTKHQIHSMERSILEKLQWYLTVPTPYVFLVR-LSSVQPDKQMENLAFFLAELTL 177
Query: 352 MHYDTLM-YCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCT 410
HY ++ Y PS IAASA+YAARCTLN+ P W+ETLK + Y EQ+++CA+LLVS +
Sbjct: 178 EHYQAIVSYSPSTIAASAIYAARCTLNRRPFWSETLKRLSSYCTEQIIECAKLLVSLQTS 357
Query: 411 VGNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQ 443
+ K ++ K+S ++G VA+LPP KN+ Q
Sbjct: 358 AADSKPNALYWKFSSEDKGFVALLPPTKNIENQ 456
>TC17019 similar to UP|Q40223 (Q40223) Cyclin, partial (21%)
Length = 652
Score = 111 bits (277), Expect = 3e-25
Identities = 57/103 (55%), Positives = 74/103 (71%), Gaps = 2/103 (1%)
Frame = +2
Query: 343 AHFLSELGMMHYDTLM-YCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCA 401
A +L+ELGMMHY + Y PS+IAASAVYAARCTL++ P W ETLK +T YSEE L +CA
Sbjct: 2 AFYLAELGMMHYPVVSSYSPSVIAASAVYAARCTLHRIPFWTETLKHYTGYSEEHLRECA 181
Query: 402 RLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVL-PPAKNLMPQ 443
+LLV+ H KL+ +++K+ +R AVA+L PAKNL Q
Sbjct: 182 KLLVNLHTAAPESKLRAIYKKFCSSDRCAVALLYVPAKNLSAQ 310
>BP030979
Length = 510
Score = 84.7 bits (208), Expect = 3e-17
Identities = 48/163 (29%), Positives = 89/163 (54%), Gaps = 9/163 (5%)
Frame = +3
Query: 127 ISPDEQIKKEKSVQK-KKEDSKKKTLTSVLTARSKAACGLTNKPKEEIVDIDASDV---- 181
+ P + KK + + KK+ + LT S A T++ + + A
Sbjct: 21 LRPQREAKKRAAAALCELRPKKKRVVXGELTNLSSPAEKPTSEKRRRVGKSKAPPPPEKR 200
Query: 182 DNELAAVEYIEDIYKFYKMVENESR----PHCYMASQPEINEKMRAILVDWLIDVHTKFE 237
D+ + Y+ DIY++ + +E + P Q ++N MR + VDWL++V +++
Sbjct: 201 DDPQMCLPYVSDIYEYLRSMEVDPSKRPLPDYVQKVQRDVNANMRGVXVDWLVEVSEEYK 380
Query: 238 LSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEE 280
L +TLY ++ +DRFL++ ++ R++LQL+G++SML+A+KYEE
Sbjct: 381 LVSDTLYFCVSYIDRFLSLNSLSRQKLQLLGVASMLVASKYEE 509
>AW164148
Length = 354
Score = 54.7 bits (130), Expect = 3e-08
Identities = 27/33 (81%), Positives = 30/33 (90%), Gaps = 1/33 (3%)
Frame = +3
Query: 168 KPKEEIVD-IDASDVDNELAAVEYIEDIYKFYK 199
KP E+IVD IDA+DV NELAAVEYI+DIYKFYK
Sbjct: 252 KPNEQIVDNIDAADVHNELAAVEYIQDIYKFYK 350
Score = 27.7 bits (60), Expect = 4.0
Identities = 16/36 (44%), Positives = 20/36 (55%)
Frame = +3
Query: 33 RNRRALGDIGNLDPVRGIEVKPNRPITRSFCAQLLA 68
RNR LGDIGN +R +++ N FC Q LA
Sbjct: 153 RNRIILGDIGNGRTLREVDINTN------FCHQPLA 242
>TC11206 similar to UP|Q84V88 (Q84V88) Cyclin D, partial (43%)
Length = 816
Score = 44.3 bits (103), Expect = 4e-05
Identities = 38/153 (24%), Positives = 65/153 (41%), Gaps = 4/153 (2%)
Frame = +2
Query: 183 NELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLET 242
N+ + + E+ + ++ NE R ++ S + R V W+ V F S T
Sbjct: 224 NDDPLILFWEEDDELVSLISNE-RGETHLGSLDLHDGGTRVEGVAWISKVSAHFGFSALT 400
Query: 243 LYLTINIVDRFLAVKTVPRRE---LQLVGISSMLMAAKYEEIWPPEVNDFVCLSDR-AYS 298
L ++ DRF+ R + QL ++ + +AAK EE P + D R +
Sbjct: 401 TVLAVSYFDRFITSPRFQRDKPWMTQLSAVACLSLAAKVEETHVPLLLDLQVEESRFVFE 580
Query: 299 HEQILVMEKIILGRLEWTLTVPTPFVFLTRFIK 331
+ I ME ++L L+W + TP F I+
Sbjct: 581 AKTIQRMELLVLSTLKWRMHPVTPISFFEHIIR 679
>AV414055
Length = 431
Score = 31.6 bits (70), Expect = 0.28
Identities = 22/86 (25%), Positives = 39/86 (44%)
Frame = +2
Query: 104 REAPKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARSKAAC 163
+E + +V P+ V E P++ + KEK ++ +KED L+ RSK
Sbjct: 65 KELVDDIAMQVAACPQVEYLVAEDVPEDIVNKEKEIEMQKED----LLSKPEQIRSKIVE 232
Query: 164 GLTNKPKEEIVDIDASDVDNELAAVE 189
G K EE+ ++ + N+ V+
Sbjct: 233 GRIRKRLEELALLEQPFIKNDKVPVK 310
>TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber
hypocotyls (Arabinogalactan protein), partial (32%)
Length = 760
Score = 31.2 bits (69), Expect = 0.36
Identities = 28/96 (29%), Positives = 36/96 (37%), Gaps = 3/96 (3%)
Frame = +1
Query: 49 GIEVKP-NRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPV--AAPGVAVAKRE 105
G V P + P+ A A A A + A P + PPAPV A+P V
Sbjct: 7 GTRVTPVSAPLASPPSKAAAAPAPVAKAPASTPPAAVPVSSPPPAPVPVASPPAPVPVSS 186
Query: 106 APKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSVQK 141
P P P EV +P + KK K +K
Sbjct: 187 PPAPAPTTPAPAVTPTAEVPAPAPSKSKKKVKKGKK 294
>TC14310 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, partial (92%)
Length = 502
Score = 31.2 bits (69), Expect = 0.36
Identities = 20/60 (33%), Positives = 27/60 (44%)
Frame = +3
Query: 93 PVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLT 152
P AP A K KP ++ + KP E ++I KE +KKK+ SKK T
Sbjct: 51 PPMAPAKAEKKPAEKKPAAEKAPAEKKPKAE-------KKIAKEGGSEKKKKKSKKSVET 209
>TC15809 weakly similar to PIR|JN0747|JN0747 histone H1-I - Volvox carteri
{Volvox carteri;}, partial (10%)
Length = 526
Score = 30.4 bits (67), Expect = 0.62
Identities = 20/56 (35%), Positives = 25/56 (43%), Gaps = 1/56 (1%)
Frame = +2
Query: 87 VAGPPAPVAAPGVAVAKREAPKPVTKRVTG-KPKPVVEVIEISPDEQIKKEKSVQK 141
+A PAP AAP K A P T + T PKP+V + SPD + K
Sbjct: 149 IAISPAPAAAP-----KSPAKTPPTPKATAPSPKPLVPTLPQSPDSSDSTPDDITK 301
>TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete
Length = 999
Score = 29.6 bits (65), Expect = 1.1
Identities = 33/115 (28%), Positives = 45/115 (38%), Gaps = 22/115 (19%)
Frame = +1
Query: 80 KKQACPNVAGPPAPVA-APGVAVAKREAPKPVTKRVTGKPK--------PVV-------- 122
++ A VA P P A P A E PK T T +PK PV
Sbjct: 184 EQPAATEVAAPEQPAAEVPAPAEPATEEPKEET---TEEPKETETETEAPVAPETQDPLE 354
Query: 123 ----EVIEISPDEQIKKE-KSVQKKKEDSKKKTLTSVLTARSKAACGLTNKPKEE 172
EV E + E++K E + ++K E+ K + + S A NKP EE
Sbjct: 355 VENKEVTEEAKPEEVKPETEKTEEKTEEPKTEEPAATTETESAAPVEEENKPAEE 519
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 29.6 bits (65), Expect = 1.1
Identities = 22/74 (29%), Positives = 32/74 (42%)
Frame = +2
Query: 68 ANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEI 127
A + A A+ K P PAP AP V+++ EAP P ++ P
Sbjct: 503 ATSPAPAPAKVKSKSPAP----APAPALAPVVSLSPSEAPGPSLSSLSPALSPAA----- 655
Query: 128 SPDEQIKKEKSVQK 141
S D ++K S+QK
Sbjct: 656 SDDSGVEKSWSMQK 697
>TC9585 similar to UP|O24078 (O24078) Protein phosphatase 2C, partial (31%)
Length = 502
Score = 29.3 bits (64), Expect = 1.4
Identities = 24/80 (30%), Positives = 34/80 (42%), Gaps = 13/80 (16%)
Frame = +3
Query: 52 VKPNRPITRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVA---------APGVAVA 102
+KPN T S CA A+A + AA ++ + + P P A +P AV
Sbjct: 189 LKPNASSTASSCASASASASSPSAAASSSPSSPFRLRLPKPPTAFSSGPSSSTSPNGAVL 368
Query: 103 KREAPK----PVTKRVTGKP 118
KR+ P PV+ G P
Sbjct: 369 KRKRPTRLDIPVSSLTFGVP 428
>BP036039
Length = 436
Score = 28.5 bits (62), Expect = 2.3
Identities = 12/39 (30%), Positives = 21/39 (53%)
Frame = +3
Query: 91 PAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEISP 129
P P A P + VAK + K ++ + K ++++I SP
Sbjct: 117 PGPTACPSLIVAKLRSTKRLSHPINSKMNHIIQIILHSP 233
>TC14481 similar to UP|PSBY_SPIOL (P80470) Photosystem II core complex
proteins psbY, chloroplast precursor (L-arginine
metabolising enzyme) (L-AME) [Contains: Photosystem II
protein psbY-1 (psbY-A1); Photosystem II protein psbY-2
(psbY-A2)], partial (50%)
Length = 796
Score = 28.5 bits (62), Expect = 2.3
Identities = 17/68 (25%), Positives = 28/68 (41%), Gaps = 5/68 (7%)
Frame = -1
Query: 60 RSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAA-----PGVAVAKREAPKPVTKRV 114
R+ C L N+ +A ++CP ++ PPA +A P A P+P
Sbjct: 655 RAGCKML--NSTQPIAGATTNSRSCPLLSLPPAAASAIIPNSPADASGAHNIPEPAANPP 482
Query: 115 TGKPKPVV 122
+ P P +
Sbjct: 481 SPSPSPTI 458
>BP063458
Length = 391
Score = 27.7 bits (60), Expect = 4.0
Identities = 10/21 (47%), Positives = 17/21 (80%)
Frame = -3
Query: 356 TLMYCPSMIAASAVYAARCTL 376
+L+ P+M+++S VYA RCT+
Sbjct: 320 SLVVIPAMLSSSPVYATRCTI 258
>TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%)
Length = 1175
Score = 27.7 bits (60), Expect = 4.0
Identities = 21/82 (25%), Positives = 31/82 (37%), Gaps = 2/82 (2%)
Frame = +2
Query: 90 PPAPVAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKK 149
PP P GVA KR + T G K + + +Q KK S+ K
Sbjct: 80 PPQPPLPSGVATNKRRRRRSFTTMSLGPKKSAPNDLVSTTQQQQKKSTSLSASSCLHKVA 259
Query: 150 TLTSVLTAR--SKAACGLTNKP 169
T T+ T + ++ C + P
Sbjct: 260 TTTTKTTTQTITQTPCAQNHHP 325
>TC12487 similar to UP|Q9SAL0 (Q9SAL0) T8K14.20 protein
(At1g79380/T8K14_20), partial (57%)
Length = 1091
Score = 27.7 bits (60), Expect = 4.0
Identities = 13/43 (30%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Frame = -1
Query: 370 YAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLL-VSFHCTV 411
Y + NKS + ++TLK H ++ E++ D ++ + FH T+
Sbjct: 917 YTSHLFYNKSMSNSKTLKQHDEH*REEVFDANHVMCIVFHSTI 789
>TC14326 weakly similar to UP|EXLP_TOBAC (Q03211) Pistil-specific
extensin-like protein precursor (PELP), partial (11%)
Length = 630
Score = 27.7 bits (60), Expect = 4.0
Identities = 15/42 (35%), Positives = 21/42 (49%), Gaps = 1/42 (2%)
Frame = +2
Query: 83 ACPNVAGPPAPVAAPGVAVAKREAP-KPVTKRVTGKPKPVVE 123
A P+V PP+PV P + P P+ K T P+P V+
Sbjct: 125 AIPSVPAPPSPVKPPTPSTPAVTVPTPPLVKAPTPPPQPPVK 250
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,142,622
Number of Sequences: 28460
Number of extensions: 94987
Number of successful extensions: 606
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 556
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 581
length of query: 447
length of database: 4,897,600
effective HSP length: 93
effective length of query: 354
effective length of database: 2,250,820
effective search space: 796790280
effective search space used: 796790280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0269a.2


