

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0266.20
(284 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
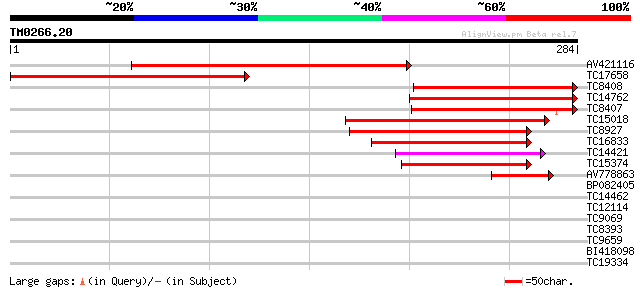
Score E
Sequences producing significant alignments: (bits) Value
AV421116 271 7e-74
TC17658 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prot... 237 1e-63
TC8408 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 160 2e-40
TC14762 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prot... 150 3e-37
TC8407 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 145 1e-35
TC15018 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, c... 104 2e-23
TC8927 similar to UP|Q9LLI0 (Q9LLI0) ClpB, partial (12%) 80 4e-16
TC16833 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-li... 60 3e-10
TC14421 similar to UP|Q39889 (Q39889) Heat shock protein , parti... 56 8e-09
TC15374 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-li... 54 3e-08
AV778863 49 7e-07
BP082405 29 1.0
TC14462 weakly similar to GB|BAA97483.1|8885553|AB025604 ripenin... 29 1.0
TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transp... 27 3.0
TC9069 27 3.0
TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenyla... 27 3.0
TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATP... 27 5.2
BI418098 27 5.2
TC19334 similar to UP|Q9YKL1 (Q9YKL1) ORF1 protein, partial (4%) 26 8.8
>AV421116
Length = 421
Score = 271 bits (694), Expect = 7e-74
Identities = 138/140 (98%), Positives = 140/140 (99%)
Frame = +1
Query: 62 QLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV 121
+LTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV
Sbjct: 1 RLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV 180
Query: 122 GSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKL 181
GSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKL
Sbjct: 181 GSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKL 360
Query: 182 EVKEIADIMLKEVFDRLKTK 201
EVKEIADIMLKEVF+RLKTK
Sbjct: 361 EVKEIADIMLKEVFERLKTK 420
>TC17658 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (33%)
Length = 908
Score = 237 bits (605), Expect = 1e-63
Identities = 118/120 (98%), Positives = 119/120 (98%)
Frame = +1
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG
Sbjct: 547 SFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 726
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN
Sbjct: 727 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 906
>TC8408 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (9%)
Length = 498
Score = 160 bits (406), Expect = 2e-40
Identities = 82/82 (100%), Positives = 82/82 (100%)
Frame = +3
Query: 203 IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS 262
IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS
Sbjct: 3 IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS 182
Query: 263 DGKVIVLNGSSGAPESLPEALP 284
DGKVIVLNGSSGAPESLPEALP
Sbjct: 183 DGKVIVLNGSSGAPESLPEALP 248
>TC14762 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (9%)
Length = 588
Score = 150 bits (378), Expect = 3e-37
Identities = 75/84 (89%), Positives = 79/84 (93%)
Frame = +1
Query: 201 KEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDA 260
+EI+L VTERFRDRVV+EGYDPSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSVI+D
Sbjct: 1 QEIELPVTERFRDRVVDEGYDPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDV 180
Query: 261 DSDGKVIVLNGSSGAPESLPEALP 284
DSDG VIVLNGSSG PESLPEALP
Sbjct: 181 DSDGNVIVLNGSSGTPESLPEALP 252
>TC8407 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (8%)
Length = 603
Score = 145 bits (365), Expect = 1e-35
Identities = 75/85 (88%), Positives = 80/85 (93%), Gaps = 2/85 (2%)
Frame = +2
Query: 202 EIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 261
EIDLSVTERF++RVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSVI+DAD
Sbjct: 2 EIDLSVTERFKERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDAD 181
Query: 262 SDGKVIVLNGS--SGAPESLPEALP 284
SDG VIVLNGS SGAPESLPE LP
Sbjct: 182 SDGNVIVLNGSSDSGAPESLPEVLP 256
>TC15018 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, chloroplast
precursor, partial (10%)
Length = 598
Score = 104 bits (259), Expect = 2e-23
Identities = 48/102 (47%), Positives = 76/102 (74%)
Frame = +2
Query: 169 LDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARP 228
+DE++VFR L K ++ EI D++L++V R+ + IDL V+E ++ V +EGY+P+YGARP
Sbjct: 2 IDEIVVFRSLEKSQLLEILDVLLQDVKKRIMSLGIDLEVSESVKNLVCKEGYNPTYGARP 181
Query: 229 LRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 270
L+RAI L+ED ++E L G+ K+GD+V+ID D +G ++V N
Sbjct: 182 LKRAITSLIEDPLSEAFLCGKCKQGDTVLIDLDVNGNLLVTN 307
>TC8927 similar to UP|Q9LLI0 (Q9LLI0) ClpB, partial (12%)
Length = 619
Score = 80.1 bits (196), Expect = 4e-16
Identities = 38/91 (41%), Positives = 62/91 (67%)
Frame = +2
Query: 171 EMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLR 230
E IVF+ L E+ +I ++ ++ V +RLK K+IDL T+ + + G+DP++GARP++
Sbjct: 2 EYIVFQPLDSKEISKIVELQMERVKNRLKQKKIDLHYTQEALELLSVLGFDPNFGARPVK 181
Query: 231 RAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 261
R I L+E+ +A +L G+ KE DS+I+DAD
Sbjct: 182 RVIQPLVENEIAMGVLRGDFKEDDSIIVDAD 274
>TC16833 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like, partial
(10%)
Length = 510
Score = 60.5 bits (145), Expect = 3e-10
Identities = 26/80 (32%), Positives = 51/80 (63%)
Frame = +3
Query: 182 EVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSM 241
++ I + L+ V R+ +++ + VTE + GYDP+YGARP++R I + +E+ +
Sbjct: 9 QISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVENEL 188
Query: 242 AEKMLAGEIKEGDSVIIDAD 261
A+ +L GE K+ D++++D +
Sbjct: 189 AKGILRGEFKDEDTILVDTE 248
>TC14421 similar to UP|Q39889 (Q39889) Heat shock protein , partial (14%)
Length = 731
Score = 55.8 bits (133), Expect = 8e-09
Identities = 28/75 (37%), Positives = 44/75 (58%)
Frame = +1
Query: 194 VFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEG 253
V RL + I ++VT+ D ++ E YDP YGARP+RR + R + ++ ++ EI E
Sbjct: 1 VASRLAERGIAMAVTDAALDYILAESYDPVYGARPIRRWLERKVVTELSRMLIRDEIDEN 180
Query: 254 DSVIIDADSDGKVIV 268
+V IDA + G +V
Sbjct: 181 STVYIDAGTKGSELV 225
>TC15374 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like, partial
(9%)
Length = 574
Score = 53.9 bits (128), Expect = 3e-08
Identities = 23/65 (35%), Positives = 44/65 (67%)
Frame = +2
Query: 197 RLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSV 256
R+ +++ + VT+ + GYDP+YGARP++R I + +E+ +A+ +L GE K+ D++
Sbjct: 8 RIADRKMKIHVTDAAVQVLGSLGYDPNYGARPVKRVIQQNVENELAKGILRGEFKDEDTI 187
Query: 257 IIDAD 261
+ID +
Sbjct: 188 LIDTE 202
>AV778863
Length = 441
Score = 49.3 bits (116), Expect = 7e-07
Identities = 23/31 (74%), Positives = 29/31 (93%)
Frame = -3
Query: 242 AEKMLAGEIKEGDSVIIDADSDGKVIVLNGS 272
A ++L+ EIKEGDS+I+DADSDGKVIVL+GS
Sbjct: 424 AREILSREIKEGDSLIVDADSDGKVIVLHGS 332
>BP082405
Length = 402
Score = 28.9 bits (63), Expect = 1.0
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +3
Query: 3 IFSGPTGVGKSELAKTLASYY 23
+ +GPTG GKS LA LAS++
Sbjct: 288 VITGPTGSGKSRLAVDLASHF 350
>TC14462 weakly similar to GB|BAA97483.1|8885553|AB025604 ripening-related
protein-like; hydrolase-like {Arabidopsis thaliana;} ,
partial (80%)
Length = 1072
Score = 28.9 bits (63), Expect = 1.0
Identities = 29/109 (26%), Positives = 47/109 (42%), Gaps = 11/109 (10%)
Frame = +1
Query: 47 LIGSPPGYV---GYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDS 103
LI S G++ Y + + R Y +LFD + +P + L +G L +
Sbjct: 91 LISSLVGFLIEMEYEYENRFRQQAERSKYDCLLFDLDDTLYP-----LRSGLANGVLHNI 255
Query: 104 KGRTVD--------FKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDS 144
KG V+ + ++ N G+++ G R IG+D DYDE S
Sbjct: 256 KGYMVEKLGIEPSKIDDLCNLLYKNYGTTMA--GMRAIGYDFDYDEYHS 396
>TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transposase-like
{Oryza sativa (japonica cultivar-group);}, partial (8%)
Length = 488
Score = 27.3 bits (59), Expect = 3.0
Identities = 13/38 (34%), Positives = 22/38 (57%)
Frame = -2
Query: 125 VIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFR 162
+++K GRK+ F L + E + YN++ VT +FR
Sbjct: 118 LVDKHGRKVNFQLTFSESIAIYNQLPH-VTNFFAFFFR 8
>TC9069
Length = 1110
Score = 27.3 bits (59), Expect = 3.0
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 4/99 (4%)
Frame = +3
Query: 129 GGRKIGFDLDYDEKDSS-YNRI--KSLVTEELKQYFRPEFLNRLDEMIVFRQL-TKLEVK 184
G R IG+D+DYD+ SS + R+ KSL+ + R L+ I+F T V+
Sbjct: 276 GLRAIGYDIDYDDFHSSVHGRLPYKSLLKPD--HVLRGILLSLPVRKIIFTNADTAHAVR 449
Query: 185 EIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPS 223
+ + L++ F+R+ I F + +G D S
Sbjct: 450 ALRSLGLEDCFERI----ISFDTLNSFNNITPNDGKDCS 554
>TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenylated Rab
receptor 2 {Arabidopsis thaliana;}, partial (78%)
Length = 1012
Score = 27.3 bits (59), Expect = 3.0
Identities = 18/53 (33%), Positives = 26/53 (48%)
Frame = -1
Query: 227 RPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAPESL 279
RPLRR RLL + +AG D+ ++DA G V+ + GA E +
Sbjct: 202 RPLRRDSERLLMRARKAGEVAGASDPADATVVDA---GGVLETGRTGGAVEDM 53
>TC9659 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (74%)
Length = 1060
Score = 26.6 bits (57), Expect = 5.2
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +1
Query: 1 SFIFSGPTGVGKSELAKTLAS 21
+F+ GP G GKS LAK +A+
Sbjct: 568 AFLLYGPPGTGKSYLAKAVAT 630
>BI418098
Length = 478
Score = 26.6 bits (57), Expect = 5.2
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +1
Query: 6 GPTGVGKSELAKTLA 20
GPTG GK+ LAKTLA
Sbjct: 181 GPTGSGKTLLAKTLA 225
>TC19334 similar to UP|Q9YKL1 (Q9YKL1) ORF1 protein, partial (4%)
Length = 507
Score = 25.8 bits (55), Expect = 8.8
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Frame = +2
Query: 120 NVGSSVIEKGGRKIGFDLDYDEKDSSYNR--IKSLVTEELKQY 160
N+ +++ + FDLD D++DSS N +K L + +K +
Sbjct: 59 NLDDDIVDAMDVDVDFDLDDDDEDSSSNAEILKELGEDMVKHF 187
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,121,928
Number of Sequences: 28460
Number of extensions: 29757
Number of successful extensions: 150
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 149
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 149
length of query: 284
length of database: 4,897,600
effective HSP length: 89
effective length of query: 195
effective length of database: 2,364,660
effective search space: 461108700
effective search space used: 461108700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0266.20


