

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
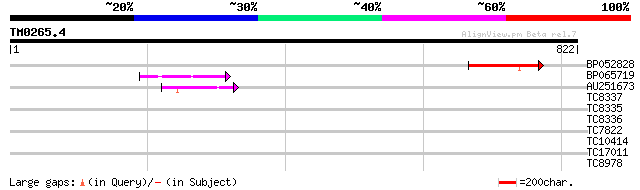
Query= TM0265.4
(822 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP052828 102 3e-22
BP065719 48 5e-06
AU251673 41 9e-04
TC8337 34 0.11
TC8335 weakly similar to UP|Q15287 (Q15287) RNA-binding protein ... 31 0.91
TC8336 weakly similar to UP|WSC2_YEAST (P53832) Cell wall integr... 31 0.91
TC7822 similar to UP|Q42139 (Q42139) H+-transporting ATP synthas... 29 2.6
TC10414 similar to GB|AAP13363.1|30023660|BT006255 At5g55710 {Ar... 29 2.6
TC17011 weakly similar to UP|Q9FMB1 (Q9FMB1) FrnE protein-like, ... 28 4.5
TC8978 28 5.9
>BP052828
Length = 523
Score = 102 bits (253), Expect = 3e-22
Identities = 50/113 (44%), Positives = 77/113 (67%), Gaps = 3/113 (2%)
Frame = +2
Query: 665 DALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVDKYVFP 724
+ + DLGA IN+MP S++ L+ G + T L +Q+A+RS P G++EDV+V+V++ +FP
Sbjct: 101 NCMLDLGAGINVMPTSIYNILHPGPLQHTGLIVQLANRSNARPKGVVEDVLVQVNELIFP 280
Query: 725 IDFVVLDMEEDAK---IPLILGRPFLATARAKIDVDKGQLILRVGKDKVRFSV 774
DF +LDME + K P+ILGRPF+ TA+ KIDVD G + + G +F++
Sbjct: 281 ADFYILDMEGETKSSRAPIILGRPFMKTAKTKIDVDNGTMSMEFGDIIAKFNI 439
>BP065719
Length = 567
Score = 48.1 bits (113), Expect = 5e-06
Identities = 34/131 (25%), Positives = 59/131 (44%)
Frame = +3
Query: 189 SQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKDAAEEWLNSQPQGSI 248
+++ G S E H+ R+ ++ E +++ FP SL A W + S+
Sbjct: 192 TKFSGDSGESTVEHIARYQIEAGDLAIN----ENLKMKYFPSSLTKNAFTWFTTLAPRSV 359
Query: 249 TTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFKKLLRKCPQHNLTQA 308
TW L F +FF R + KD+ + + E++ + RF+ L +C H +++
Sbjct: 360 HTWAQLERIFHEQFF-RGECKVSXKDLASVKRKPAESIDDYLNRFRMLKSRCFTH-VSEH 533
Query: 309 ELVATFYDGLE 319
ELV GLE
Sbjct: 534 ELVVLXAGGLE 566
>AU251673
Length = 413
Score = 40.8 bits (94), Expect = 9e-04
Identities = 34/118 (28%), Positives = 54/118 (44%), Gaps = 6/118 (5%)
Frame = +2
Query: 220 SEAIRLSLFPFSLKDAAEEWLN----SQPQGSIT-TWEDLAEKFTTRFFPRALLRKLKKD 274
S+ + L F L+ A +W N ++P GS TW D + +F RF P+++ +D
Sbjct: 8 SDTRAVELASFQLEGVARDWYNVLTRAKPVGSPPWTWADFSAEFMNRFLPQSVRDGFVRD 187
Query: 275 IMNFVQSAEENLYEAWERFKKLLRKCPQHNLTQAELVATFYDGL-EYSWRFGLDTASS 331
Q+ + E F L R P + L + E V F GL EY ++ + + SS
Sbjct: 188 FERLEQAEGMTVSEYSAHFTHLSRYVP-YPLLEEERVKRFVRGLKEYLFKSVVGSKSS 358
>TC8337
Length = 553
Score = 33.9 bits (76), Expect = 0.11
Identities = 15/40 (37%), Positives = 24/40 (59%)
Frame = +1
Query: 506 HENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIKK 545
HE +DDV + + HE +V E +DK E+K E V+++
Sbjct: 124 HEESDDVAVKDESLDHEESIRVVVEHDDKTEDKTEDVVEE 243
>TC8335 weakly similar to UP|Q15287 (Q15287) RNA-binding protein
(RNA-binding protein S1, serine-rich domain), partial
(11%)
Length = 577
Score = 30.8 bits (68), Expect = 0.91
Identities = 15/44 (34%), Positives = 24/44 (54%)
Frame = +2
Query: 506 HENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIKKRVSE 549
HE +DDV + + HE +V E +DK E+ E + K +V +
Sbjct: 143 HEESDDVAVKDEALDHEESVKVAVEHDDKTEDVVEELQKLKVDD 274
>TC8336 weakly similar to UP|WSC2_YEAST (P53832) Cell wall integrity and
stress response component 2 precursor, partial (6%)
Length = 600
Score = 30.8 bits (68), Expect = 0.91
Identities = 15/44 (34%), Positives = 24/44 (54%)
Frame = +3
Query: 506 HENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIKKRVSE 549
HE +DDV + + HE +V E +DK E+ E + K +V +
Sbjct: 186 HEESDDVAVKDEALDHEESVKVAVEHDDKTEDVVEELQKLKVDD 317
>TC7822 similar to UP|Q42139 (Q42139) H+-transporting ATP synthase CHAIN9 -
like protein (AT4G32260/F10M6_100) , partial (76%)
Length = 907
Score = 29.3 bits (64), Expect = 2.6
Identities = 13/38 (34%), Positives = 19/38 (49%)
Frame = +2
Query: 135 ITLRLCEQEVENANRSLRELTSAPMSYDYPGSIVFPEG 172
+T RLC NR R + P + D+ G + FP+G
Sbjct: 278 VTTRLCSTIPGGGNRESRAVRFQPDTPDHSGGVSFPDG 391
>TC10414 similar to GB|AAP13363.1|30023660|BT006255 At5g55710 {Arabidopsis
thaliana;}, partial (34%)
Length = 504
Score = 29.3 bits (64), Expect = 2.6
Identities = 15/43 (34%), Positives = 21/43 (47%)
Frame = -3
Query: 209 NCNTYRVSNVSSEAIRLSLFPFSLKDAAEEWLNSQPQGSITTW 251
NC Y+ N E + L + FS+ A EEW+ +G T W
Sbjct: 430 NCR-YQWLNEDLERVELGDYVFSVLYAVEEWVEVAERGDQTVW 305
>TC17011 weakly similar to UP|Q9FMB1 (Q9FMB1) FrnE protein-like, partial
(91%)
Length = 1033
Score = 28.5 bits (62), Expect = 4.5
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = -3
Query: 276 MNFVQSAEENLYEAWERFKKLLRKCPQHNL 305
+NF + AE L AW+ KK R CP +L
Sbjct: 722 INFWEGAEIQLVAAWKALKKTSRGCPPLSL 633
>TC8978
Length = 506
Score = 28.1 bits (61), Expect = 5.9
Identities = 13/48 (27%), Positives = 24/48 (49%)
Frame = -1
Query: 449 KKSLEELLESFINRMESNYKIQDAAIKNLKSQFGQLAKQMAKIPQGKF 496
KK E+L + R NY+ + ++K + ++ Q+AKI G +
Sbjct: 284 KKHSNEVLSRIVARSPKNYQYSQVYLDSIKVKSTKVLSQVAKITGGSY 141
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.139 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,052,348
Number of Sequences: 28460
Number of extensions: 153876
Number of successful extensions: 974
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 962
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 972
length of query: 822
length of database: 4,897,600
effective HSP length: 98
effective length of query: 724
effective length of database: 2,108,520
effective search space: 1526568480
effective search space used: 1526568480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0265.4


