

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0263.11
(526 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
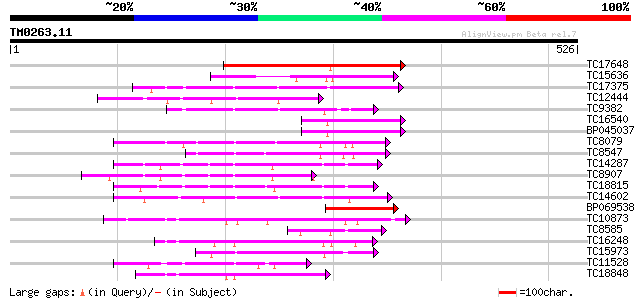
Score E
Sequences producing significant alignments: (bits) Value
TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%) 178 2e-45
TC15636 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial... 146 7e-36
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 144 5e-35
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 94 4e-20
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 94 4e-20
TC16540 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial... 94 7e-20
BP045037 94 7e-20
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 88 3e-18
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 86 1e-17
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 84 4e-17
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 76 2e-14
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 75 3e-14
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 74 4e-14
BP069538 74 6e-14
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 71 5e-13
TC8585 weakly similar to PIR|T51339|T51339 mitogen-activated pro... 70 8e-13
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 70 8e-13
TC15973 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F... 69 1e-12
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 69 2e-12
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 69 2e-12
>TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%)
Length = 741
Score = 178 bits (451), Expect = 2e-45
Identities = 89/175 (50%), Positives = 114/175 (64%), Gaps = 6/175 (3%)
Frame = +3
Query: 199 LHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMS 258
L GL YLH RH++HRD KP++LLVN +GE KITDFG+SA L +S+ TFVGT YMS
Sbjct: 3 LQGLVYLHNERHVIHRDSKPSHLLVNHQGEVKITDFGVSAMLATSMGQRDTFVGTYNYMS 182
Query: 259 PERIRNESYSYPADIWSLGLALLECGTGEFPYTANEGP------VNLMLQILDDPSPSPS 312
P RI +Y Y +DIWSLG+ +LEC G FPY +E L+ I++ P PS
Sbjct: 183 PARISGSTYDYSSDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLAAIVESPPPSAP 362
Query: 313 KQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETVEVDLAGYVRSVFDP 367
FSPEFCSF+ +C+QKDP R T+ +LL HPFI K+E ++DL V S+ P
Sbjct: 363 SDQFSPEFCSFVSSCIQKDPQDRLTSLELLDHPFIKKFEDKDLDLEILVGSLEPP 527
>TC15636 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (46%)
Length = 1184
Score = 146 bits (369), Expect = 7e-36
Identities = 81/182 (44%), Positives = 108/182 (58%), Gaps = 8/182 (4%)
Frame = +2
Query: 187 PEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAM 246
PEP L+S+ +++L GL YLH +H++HRD+KP+NLL+N +GEP
Sbjct: 2 PEPYLASICKQVLKGLMYLHHEKHIIHRDLKPSNLLINHRGEP----------------- 130
Query: 247 CATFVGTVTYMSPERIRN--ESYSYPADIWSLGLALLECGTGEFPYTA---NEGPVN--- 298
ERI + Y+Y +DIWSLGL L C TG FPYT +EG N
Sbjct: 131 -------------ERINGSQDGYNYKSDIWSLGLMFLRCATGMFPYTPPDQSEGWENIYQ 271
Query: 299 LMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETVEVDLA 358
L+ I+++PSPS S FS EFCSFI ACLQK+P RP+A +L+ HPFI Y+ + VDL+
Sbjct: 272 LIEAIVENPSPSVSSDEFSSEFCSFIAACLQKNPKDRPSAPELMRHPFINMYDDLAVDLS 451
Query: 359 GY 360
Y
Sbjct: 452 AY 457
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 144 bits (362), Expect = 5e-35
Identities = 90/257 (35%), Positives = 149/257 (57%), Gaps = 6/257 (2%)
Frame = +3
Query: 115 RIIALKKINIFEKEKR-----QQLLTEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALE 169
++ A+K++ +F +K +QL EI L + + +V+++G+ +S +S+ LE
Sbjct: 3 QMCAIKEVKVFSDDKTSKECLKQLNQEINLLNQFS-HPNIVQYYGSELGEES--LSVYLE 173
Query: 170 YMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEP 229
Y+ GGS+ +L+++ EP++ + ++++ GL+YLH R+ VHRDIK AN+LV+ GE
Sbjct: 174 YVSGGSIHKLLQEYGAFKEPVIQNYTRQIVSGLAYLHS-RNTVHRDIKGANILVDPNGEI 350
Query: 230 KITDFGISAGLESSVAMCATFVGTVTYMSPERIRNES-YSYPADIWSLGLALLECGTGEF 288
K+ DFG+S + S+ +M + F G+ +M+PE + N + Y P DI SLG +LE T +
Sbjct: 351 KLADFGMSKHINSAASMLS-FKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKP 527
Query: 289 PYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFIT 348
P++ EG V + +I + + S + +FI CLQ+DP RPTA+ LL HPFI
Sbjct: 528 PWSQFEG-VAAIFKIGNSKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIR 704
Query: 349 KYETVEVDLAGYVRSVF 365
+V A R F
Sbjct: 705 DQSATKVANASITRDAF 755
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 94.4 bits (233), Expect = 4e-20
Identities = 61/219 (27%), Positives = 113/219 (50%), Gaps = 9/219 (4%)
Frame = +2
Query: 82 KTYRCGSREMRIFGAIGSGASSVVQRAIHIPKHRIIALKKINIFEKEKRQQLLTEIRTLC 141
+TY S + + +G GAS+ V RA+ +P+ +A+K +++ ++ L +IR
Sbjct: 62 RTYSLNSGDYNLLEEVGYGASATVYRAVFLPRDEEVAVKCVDL---DRCNANLEDIRREA 232
Query: 142 EAPC---YQGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRR--IPEPILSSMFQ 196
+ ++ +V H +F ++ + + +M GS +++ E + S+ +
Sbjct: 233 QTMSLIDHRNVVRAHWSFVV--DRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSILK 406
Query: 197 KLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCA---TFVGT 253
+ L L YLH H +HRD+K N+L+ G+ K+ DFG+SA + + TFVGT
Sbjct: 407 ETLKALEYLHRHGH-IHRDVKAGNILLGSDGQVKLADFGVSASMFDAGERQRNRNTFVGT 583
Query: 254 VTYMSPERIR-NESYSYPADIWSLGLALLECGTGEFPYT 291
+M+PE ++ Y++ AD+WS G+ LE G P++
Sbjct: 584 PCWMAPEVLQPGTGYNFKADVWSFGITALELAHGHAPFS 700
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 94.4 bits (233), Expect = 4e-20
Identities = 68/202 (33%), Positives = 108/202 (52%), Gaps = 5/202 (2%)
Frame = +2
Query: 146 YQGLVEFHGAFY-TPDSGQISIALEYMDGGSLADILRKHRRIPE-PILSSMFQKLLHGLS 203
++ +V+F GA TP+ + I E+M GSL D L K R + + P L + + G++
Sbjct: 107 HKNVVQFIGACTRTPN---LCIVTEFMSRGSLYDFLHKQRGVFKLPSLLKVAIDVSKGMN 277
Query: 204 YLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIR 263
YLH +++HRD+K NLL++ K+ DFG++ + S M A GT +M+PE I
Sbjct: 278 YLHQ-NNIIHRDLKTGNLLMDENELVKVADFGVARVITQSGVMTAE-TGTYRWMAPEVIE 451
Query: 264 NESYSYPADIWSLGLALLECGTGEFPY---TANEGPVNLMLQILDDPSPSPSKQTFSPEF 320
++ Y AD++S G+AL E TGE PY T + V ++ + L PS K T P
Sbjct: 452 HKPYDQKADVFSFGIALWELLTGELPYSYLTPLQAAVGVVQKGL---RPSIPKNT-HPRL 619
Query: 321 CSFIDACLQKDPDVRPTAEQLL 342
+ C ++DP RP +++
Sbjct: 620 SELLQRCWKQDPIERPAFSEII 685
>TC16540 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (30%)
Length = 672
Score = 93.6 bits (231), Expect = 7e-20
Identities = 47/103 (45%), Positives = 62/103 (59%), Gaps = 6/103 (5%)
Frame = +1
Query: 271 ADIWSLGLALLECGTGEFPYTAN------EGPVNLMLQILDDPSPSPSKQTFSPEFCSFI 324
+DIWSLGL LLEC G FPY E L+ ++D P PS + FS EFCSFI
Sbjct: 1 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 180
Query: 325 DACLQKDPDVRPTAEQLLFHPFITKYETVEVDLAGYVRSVFDP 367
ACLQKDP R +A++L+ PF + Y+ ++VDL+ Y + P
Sbjct: 181 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSP 309
>BP045037
Length = 565
Score = 93.6 bits (231), Expect = 7e-20
Identities = 47/103 (45%), Positives = 62/103 (59%), Gaps = 6/103 (5%)
Frame = -2
Query: 271 ADIWSLGLALLECGTGEFPYTAN------EGPVNLMLQILDDPSPSPSKQTFSPEFCSFI 324
+DIWSLGL LLEC G FPY E L+ ++D P PS + FS EFCSFI
Sbjct: 564 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 385
Query: 325 DACLQKDPDVRPTAEQLLFHPFITKYETVEVDLAGYVRSVFDP 367
ACLQKDP R +A++L+ PF + Y+ ++VDL+ Y + P
Sbjct: 384 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSP 256
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 88.2 bits (217), Expect = 3e-18
Identities = 78/291 (26%), Positives = 131/291 (44%), Gaps = 34/291 (11%)
Frame = +3
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKI-NIFEKEK-RQQLLTEIRTLCEAPCYQGLVEFHG 154
IG GA +V ++ + ++A+KKI N F+ ++ L EI+ L ++ ++
Sbjct: 309 IGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLD-HENVIALRD 485
Query: 155 AFYTP---DSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHL 211
P + + I E MD L I+R ++ + E ++L GL Y+H ++
Sbjct: 486 VIPPPLRREFTDVYITTELMDT-DLHQIIRSNQGLSEEHCQYFLYQVLRGLKYIHSA-NI 659
Query: 212 VHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNES-YSYP 270
+HRD+KP+NLL+N + KI DFG++ + M +V T Y +PE + N S Y+
Sbjct: 660 IHRDLKPSNLLLNSNCDLKIIDFGLARPTVENDFM-TEYVVTRWYRAPELLLNSSDYTSA 836
Query: 271 ADIWSLGLALLECGTGE--FPYTANEGPVNLMLQILDDPSPS------------------ 310
D+WS+G +E + FP + + L+ ++L P+ +
Sbjct: 837 IDVWSVGCIFMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIRQLP 1016
Query: 311 -----PSKQTF---SPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETV 353
P + F P +D L DP R T EQ L HP++ K V
Sbjct: 1017QYPRQPLTKVFPHVHPMAMDLVDKMLTIDPTKRITVEQALAHPYLEKLHDV 1169
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 85.9 bits (211), Expect = 1e-17
Identities = 63/218 (28%), Positives = 103/218 (46%), Gaps = 28/218 (12%)
Frame = +1
Query: 164 ISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLV 223
+ IA E MD L I+R ++ + E ++L GL Y+H +++HRD+KP+NLL+
Sbjct: 64 VYIAYELMDT-DLHQIIRSNQGLSEEHCQYFLYQILRGLKYIHSA-NVLHRDLKPSNLLL 237
Query: 224 NLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNES-YSYPADIWSLGLALLE 282
N + KI DFG+ A + S +V T Y +PE + N S Y+ D+WS+G +E
Sbjct: 238 NANCDLKICDFGL-ARVTSETDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFME 414
Query: 283 CGTGE--FPYTANEGPVNLMLQILDDPS------------------PSPSKQTFS----- 317
+ FP + + L+++++ PS P +Q+F
Sbjct: 415 LMDRKPLFPGRDHVHQLRLLMELIGTPSEDDLGFLNENAKRYIRQLPPYRRQSFQEKFPQ 594
Query: 318 --PEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETV 353
PE ++ L DP R T E+ L HP++T +
Sbjct: 595 VHPEAIDLVEKMLTFDPRKRITVEEALAHPYLTSLHDI 708
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 84.3 bits (207), Expect = 4e-17
Identities = 70/256 (27%), Positives = 125/256 (48%), Gaps = 6/256 (2%)
Frame = +3
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIFEKEKRQQLLTEIR---TLCEAPCYQGLVEFH 153
+G G + V ++ + +A+K I EK K+ +L+ +I+ ++ + +VE
Sbjct: 450 LGQGNFAKVYHGRNLATNENVAIKVIKK-EKLKKDRLVKQIKREVSVMRLVRHPHIVELK 626
Query: 154 GAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVH 213
T G+I + +EY+ GG L + K + + E FQ+L+ + + H R + H
Sbjct: 627 EVMAT--KGKIFLVMEYVKGGELFTKVNKGK-LNEDDARKYFQQLISAVDFCHS-RGVTH 794
Query: 214 RDIKPANLLVNLKGEPKITDFGISAGLES--SVAMCATFVGTVTYMSPERIRNESY-SYP 270
RD+KP NLL++ + K++DFG+SA E M T GT Y++PE ++ + Y
Sbjct: 795 RDLKPENLLLDENEDLKVSDFGLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKKGYDGSK 974
Query: 271 ADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQK 330
ADIWS G+ L +G P+ E + + + P + SP+ + I L
Sbjct: 975 ADIWSCGVILYALLSGYLPF-QGENVMRIYRKAFKAEYEFP--EWISPQAKNLISNLLVA 1145
Query: 331 DPDVRPTAEQLLFHPF 346
DP+ R + +++ P+
Sbjct: 1146DPEKRYSIPEIISDPW 1193
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 75.9 bits (185), Expect = 2e-14
Identities = 66/231 (28%), Positives = 115/231 (49%), Gaps = 13/231 (5%)
Frame = +3
Query: 67 SSRSVDESHDSDHSEKTYRCGSREM-----RIFGAIGSGASSVVQRAIHIPKHRIIALKK 121
++R+ + D +T R R + I +G G + V ++ + +A+K
Sbjct: 255 TTRNKTINMDQRQQSRTMRAAKRNILFNKYEIGKILGQGNFAKVYHGRNMETNESVAIKV 434
Query: 122 INIFEKEKRQQLLTEIR---TLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLAD 178
I E+ K+++L+ +I+ ++ + +VE T +I + +EY+ GG L
Sbjct: 435 IKK-ERLKKERLVKQIKREVSVMRLVRHPHIVELKEVMATKT--KIFMVVEYVKGGELFA 605
Query: 179 ILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISA 238
L K + + E FQ+L+ + + H R + HRD+KP NLL++ + K++DFG+S+
Sbjct: 606 KLTKGK-MTEVAARKYFQQLISAVDFCHS-RGVTHRDLKPENLLLDDNEDLKVSDFGLSS 779
Query: 239 GLES--SVAMCATFVGTVTYMSPERIRNESY-SYPADIWSLGLAL--LECG 284
E S M T GT Y++PE ++ + Y ADIWS G+ L L CG
Sbjct: 780 LPEQRRSDGMLLTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLCG 932
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 74.7 bits (182), Expect = 3e-14
Identities = 70/253 (27%), Positives = 116/253 (45%), Gaps = 7/253 (2%)
Frame = +2
Query: 97 IGSGASSVVQRAIHIPKHRIIALK---KINIFEKEKRQQLLTEIRTLCEAPCYQGLVEFH 153
+G G + V A I +A+K K I ++ Q+ EI ++ + +V
Sbjct: 56 LGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREI-SIMRLVRHPNIVNLK 232
Query: 154 GAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVH 213
T +I +EY+ GG L + K + + + + FQ+L+ + Y H R + H
Sbjct: 233 EVMATKT--KIFFIMEYIRGGELFAKVAKGK-LKDDLARRYFQQLISAVDYCHS-RGVSH 400
Query: 214 RDIKPANLLVNLKGEPKITDFGISAGLESSV---AMCATFVGTVTYMSPERIRNESY-SY 269
RD+KP NLL++ K++DFG+S GL + + T GT Y++PE +R + Y +
Sbjct: 401 RDLKPENLLLDENENLKVSDFGLS-GLPEQLRQDGLLHTQCGTPAYVAPEVLRKKGYDGF 577
Query: 270 PADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQ 329
D WS G+ L G P+ +E + + ++L P FSPE I L
Sbjct: 578 KTDTWSCGVILYALLAGCLPF-QHENLMTMYNKVLRAEFQFP--PWFSPESKKLISKILV 748
Query: 330 KDPDVRPTAEQLL 342
DP+ R T ++
Sbjct: 749 ADPNRRITISSIM 787
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 74.3 bits (181), Expect = 4e-14
Identities = 67/267 (25%), Positives = 117/267 (43%), Gaps = 8/267 (2%)
Frame = +2
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKIN---IFEKEKRQQLLTEIRTLCEAPCYQGLVEFH 153
IG G + R H A+K I+ + + R L E + + + +++
Sbjct: 86 IGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSPHPNILQIF 265
Query: 154 GAFYTPDSGQISIALEYMDGGSLAD--ILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHL 211
F D +S+ +E +L D + IPE + + ++LL +++ H + +
Sbjct: 266 DVF--EDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHCHRLG-V 436
Query: 212 VHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESYSYPA 271
HRD+KP N+L G+ K+ DFG + M VGT Y++PE + Y
Sbjct: 437 AHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGRRMSGV-VGTPYYVAPEVLMGREYGEKV 613
Query: 272 DIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPSPSK--QTFSPEFCSFIDACLQ 329
D+WS G+ L +G P+ + + ++ PS+ + SP + +
Sbjct: 614 DVWSCGVILYIMLSGTPPF-YGDSAAEIFEAVIRGNLRFPSRIFRNVSPAAKDLLRKMIC 790
Query: 330 KDPDVRPTAEQLLFHP-FITKYETVEV 355
+DP R +AEQ L HP F++ +TV V
Sbjct: 791 RDPSNRISAEQALRHPWFLSAGDTVNV 871
>BP069538
Length = 532
Score = 73.9 bits (180), Expect = 6e-14
Identities = 35/67 (52%), Positives = 46/67 (68%)
Frame = -3
Query: 294 EGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITKYETV 353
E L+ I+++PSPS S FS EFCSFI ACLQK+P RP+A +L+ HPFI Y+ +
Sbjct: 530 ENIYQLIEAIVENPSPSVSSDEFSSEFCSFIAACLQKNPKDRPSAPELMRHPFINMYDDL 351
Query: 354 EVDLAGY 360
VDL+ Y
Sbjct: 350 AVDLSAY 330
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 70.9 bits (172), Expect = 5e-13
Identities = 79/309 (25%), Positives = 130/309 (41%), Gaps = 24/309 (7%)
Frame = +1
Query: 88 SREMRIFGAIGSGASSVVQRAIHIPKHRIIALKKINIFEKEKRQQLLTEIRTLCEAPCYQ 147
+R + IG G V + + +A+K+++ ++ Q+ + E+ L +
Sbjct: 442 TRNFKEANLIGEGGFGKVYKG-RLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLH-HT 615
Query: 148 GLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLL----HGLS 203
LV G Y D Q + EYM GSL D L + EP+ S K+ GL
Sbjct: 616 NLVRLIG--YCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLE 789
Query: 204 YLHGVRH--LVHRDIKPANLLVNLKGEPKITDFGIS--AGLESSVAMCATFVGTVTYMSP 259
YLH +++RD+K AN+L++ + PK++DFG++ + + + +GT Y +P
Sbjct: 790 YLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAP 969
Query: 260 ERIRNESYSYPADIWSLGLALLECGTGEFPYTANEGPVNLMLQILDDPSPS--------- 310
E + + +DI+S G+ LLE TG + P L P S
Sbjct: 970 EYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMV 1149
Query: 311 -PSKQTFSPEFC-----SFIDACLQKDPDVRPTAEQLLFH-PFITKYETVEVDLAGYVRS 363
P Q P C + CLQ+ P RP ++ ++ + E A Y+
Sbjct: 1150DPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQGSPE---AHYLYG 1320
Query: 364 VFDPTQRMK 372
V P RM+
Sbjct: 1321VQQPPSRME 1347
>TC8585 weakly similar to PIR|T51339|T51339 mitogen-activated protein
kinase kinase 4 [validated] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(24%)
Length = 549
Score = 70.1 bits (170), Expect = 8e-13
Identities = 47/101 (46%), Positives = 57/101 (55%), Gaps = 9/101 (8%)
Frame = +2
Query: 258 SPERIRNESYS-----YPADIWSLGLALLECGTGEFPYT-ANEGP--VNLMLQI-LDDPS 308
SPER E+ + ADIWSLGL L E G FP+ A + P V LM I DP
Sbjct: 2 SPERFDPEASGGNYNGFAADIWSLGLTLFELYVGHFPFLQAGQRPDWVTLMCGICFGDPP 181
Query: 309 PSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLLFHPFITK 349
P +T SPEF SF++ CL+K+ R TA QLL HPF+ K
Sbjct: 182 SLP--ETASPEFRSFVECCLKKESGERWTAAQLLAHPFVCK 298
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 70.1 bits (170), Expect = 8e-13
Identities = 59/224 (26%), Positives = 107/224 (47%), Gaps = 17/224 (7%)
Frame = +2
Query: 135 TEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRKHRRIPE---PIL 191
+EI+TL E ++ +++ HG + S + E+++GGSL +L +
Sbjct: 2 SEIQTLTEIR-HRNVIKLHG--FCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKR 172
Query: 192 SSMFQKLLHGLSYLHG--VRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSVAMCAT 249
++ + + + LSY+H ++HRDI N+L++L+ E ++DFG + L+
Sbjct: 173 VNVVKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFLKPGSHTWTA 352
Query: 250 FVGTVTYMSPERIRNESYSYPADIWSLGLALLECGTGEFP-------YTANEGPV--NLM 300
F GT Y +PE + + D++S G+ LE G P + + P+ +L+
Sbjct: 353 FAGTFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDLL 532
Query: 301 LQILDDPSPSPSKQTFSPE---FCSFIDACLQKDPDVRPTAEQL 341
L + D P ++ E S ACL+K+P RPT +Q+
Sbjct: 533 LIDILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQV 664
>TC15973 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F4P13_4
{Arabidopsis thaliana;}, partial (49%)
Length = 944
Score = 69.3 bits (168), Expect = 1e-12
Identities = 50/175 (28%), Positives = 84/175 (47%), Gaps = 5/175 (2%)
Frame = +3
Query: 173 GGSLADILRKHRR--IPEPILSSMFQKLLHGLSYLHGVRHLVHRDIKPANLLVNLKGEPK 230
GG+L L K+RR + ++ + L GLSYLH + +VHRD+K N+L++ K
Sbjct: 3 GGTLKSYLIKNRRRKLAFKVVIQLALDLARGLSYLHSQK-IVHRDVKTENMLLDKTRTVK 179
Query: 231 ITDFGISAGLESSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLECGTGEFPY 290
I DFG++ S+ GT+ YM+PE + Y+ D++S G+ L E + PY
Sbjct: 180 IADFGVARVEASNPNDMTGETGTLGYMAPEVLNGNPYNRKCDVYSFGICLWEIYCCDMPY 359
Query: 291 ---TANEGPVNLMLQILDDPSPSPSKQTFSPEFCSFIDACLQKDPDVRPTAEQLL 342
+ +E ++ Q L P + + + C PD RP ++++
Sbjct: 360 PDLSFSEITSAVVRQNLRPEIP----RCCPSSLANVMKKCWDATPDKRPEMDEVV 512
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 68.9 bits (167), Expect = 2e-12
Identities = 59/191 (30%), Positives = 94/191 (48%), Gaps = 7/191 (3%)
Frame = +3
Query: 97 IGSGASSVVQRAIHIPKHRIIALKKINIFEK--EKRQQLLTEIRTLCEAPCYQGLVEFHG 154
+GSG V + A ++A+K I +K E Q+ + R+L + ++ F
Sbjct: 96 LGSGNFGVARLARDKNTGELVAVKYIERGKKIDENVQREIINHRSLR----HPNIIRFKE 263
Query: 155 AFYTPDSGQISIALEYMDGGSLADILRKHRRIPEPILSSMFQKLLHGLSYLHGVRHLVHR 214
TP ++I LEY GG L + + R E FQ+L+ G+SY H + + HR
Sbjct: 264 VLLTPT--HLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSM-EICHR 434
Query: 215 DIKPANLLVNLKGEP--KITDFGISAGLESSV--AMCATFVGTVTYMSPERIRNESY-SY 269
D+K N L++ P KI DFG S +S++ + + VGT Y++PE + + Y
Sbjct: 435 DLKLENTLLDGNPSPRLKICDFGYS---KSAILHSQPKSTVGTPAYIAPEVLSRKEYDGK 605
Query: 270 PADIWSLGLAL 280
AD+WS G+ L
Sbjct: 606 VADVWSCGVTL 638
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 68.9 bits (167), Expect = 2e-12
Identities = 55/188 (29%), Positives = 83/188 (43%), Gaps = 7/188 (3%)
Frame = +2
Query: 117 IALKKINIFEKEKRQQLLTEIRTLCEAPCYQGLVEFHGAFYTPDSGQISIALEYMDGGSL 176
IA+KK+ + + E+ L ++ L+ G Y Q I +YM SL
Sbjct: 377 IAVKKLKAMNSKAEMEFAVEVEVLGRVR-HKNLLGLRG--YCVGDDQRLIVYDYMPNLSL 547
Query: 177 ADILRKHRRIPEPILSSMFQKLL----HGLSYLHG--VRHLVHRDIKPANLLVNLKGEPK 230
L + + K+ G+ YLH H++HRDIK +N+L+N EP
Sbjct: 548 LSHLHGQFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPL 727
Query: 231 ITDFGISAGLESSVAMCATFV-GTVTYMSPERIRNESYSYPADIWSLGLALLECGTGEFP 289
+ DFG + + V+ T V GT+ Y++PE S D++S G+ LLE TG P
Sbjct: 728 VADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKP 907
Query: 290 YTANEGPV 297
G V
Sbjct: 908 IEKLPGGV 931
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,450,676
Number of Sequences: 28460
Number of extensions: 139445
Number of successful extensions: 890
Number of sequences better than 10.0: 196
Number of HSP's better than 10.0 without gapping: 826
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 834
length of query: 526
length of database: 4,897,600
effective HSP length: 94
effective length of query: 432
effective length of database: 2,222,360
effective search space: 960059520
effective search space used: 960059520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0263.11


