

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.7
(320 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
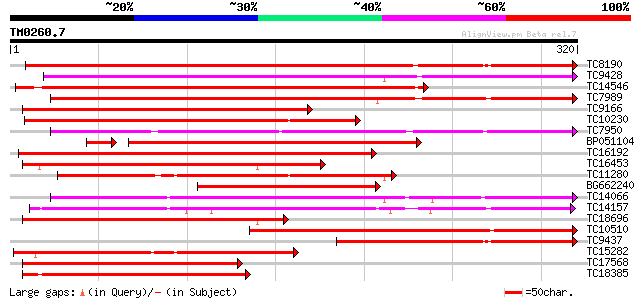
Score E
Sequences producing significant alignments: (bits) Value
TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 p... 390 e-109
TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), pa... 248 7e-67
TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxida... 234 1e-62
TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , par... 230 3e-61
TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%) 194 2e-50
TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precurso... 192 5e-50
TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), part... 191 1e-49
BP051104 188 2e-49
TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%) 188 1e-48
TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, part... 183 4e-47
TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), p... 182 7e-47
BG662240 179 4e-46
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 179 4e-46
TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , p... 172 5e-44
TC18696 similar to UP|Q40372 (Q40372) Peroxidase precursor, part... 169 4e-43
TC10510 similar to PIR|B56555|B56555 peroxidase , anionic, precu... 155 7e-39
TC9437 similar to UP|Q7XYR7 (Q7XYR7) Class III peroxidase, parti... 150 4e-37
TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fra... 144 3e-35
TC17568 similar to UP|Q8RVP6 (Q8RVP6) Gaiacol peroxidase , part... 134 2e-32
TC18385 similar to UP|Q9XFL6 (Q9XFL6) Peroxidase 5, partial (38%) 134 3e-32
>TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (96%)
Length = 1202
Score = 390 bits (1003), Expect = e-109
Identities = 198/311 (63%), Positives = 238/311 (75%)
Frame = +1
Query: 10 FLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQ 69
F CL+G SA LS+ FY +TCP L TI++ V AV+KE RMGASLLRLHFHDCFVQ
Sbjct: 100 FFFLFCLIGIGSAQLSSTFYAKTCPLVLATIKTQVNLAVAKEARMGASLLRLHFHDCFVQ 279
Query: 70 GCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARD 129
GCDAS+LLDDT SFTGEK + PNANS+RG++VID IK ++ES+CPGVVSCADI+ VAARD
Sbjct: 280 GCDASILLDDTSSFTGEKTAGPNANSVRGYDVIDTIKSKVESLCPGVVSCADIVAVAARD 459
Query: 130 SVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSG 189
SV ALGG W + LGRRDSTTASL ++NS+LP P +L GL +A+ KGF EMV LSG
Sbjct: 460 SVVALGGFSWAVPLGRRDSTTASLSSANSELPGPSSNLDGLNTAFSNKGFTTREMVALSG 639
Query: 190 AHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAF 249
+HTIG +RC+F+R RIY ET+ID +FA ++Q NC F+GG D+N SP D+T+ FDN +
Sbjct: 640 SHTIGQARCLFFRTRIYYETHIDSTFAKNLQGNCPFNGG--DSNLSPLDTTSPTTFDNGY 813
Query: 250 YQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSD 309
Y+NL +QKGL HSDQ L+ NG G+TDSQV SY N F DFANAMVKM LSPLTGS
Sbjct: 814 YRNLQSQKGLFHSDQVLF-NG-GSTDSQVNSYVTNPASFKTDFANAMVKMGNLSPLTGSS 987
Query: 310 GQIRKNCRVVN 320
GQIR +CR N
Sbjct: 988 GQIRTHCRKTN 1020
>TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1354
Score = 248 bits (634), Expect = 7e-67
Identities = 129/308 (41%), Positives = 181/308 (57%), Gaps = 7/308 (2%)
Frame = +2
Query: 20 TSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDD 79
+ A L FY +TCP +R V +PRM SL+RLHFHDCFVQGCDAS+LL+D
Sbjct: 104 SDAQLDNSFYKDTCPNVHSIVREVVRNVSKSDPRMLGSLIRLHFHDCFVQGCDASILLND 283
Query: 80 TDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRW 139
T + E+ +FPN NS+RG +V++ IK +E+ CP VSCADIL +AA S G W
Sbjct: 284 TSTIVSEQGAFPNNNSIRGLDVVNQIKTAVENACPNTVSCADILALAAEISSVLANGVDW 463
Query: 140 DLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCI 199
+ LGRRDS TA+ +N +LP P +LS L +++ + N T++V LSGAHTIG +C
Sbjct: 464 KVPLGRRDSLTANRSLANQNLPGPNFNLSQLNASFAVQNLNITDLVALSGAHTIGRGQCQ 643
Query: 200 FYRDRIYNETN-------IDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQN 252
F+ +R+YN +N ++ ++ +++ C G T + D TT D+ ++ N
Sbjct: 644 FFVNRLYNFSNTGNPDPTLNTTYLQTLRSICPHGGPGT--TLAHLDPTTPDTLDSQYFSN 817
Query: 253 LLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQI 312
L KGL SD L+ T + V S+S N F E+F +M+KMS + LTGS G+I
Sbjct: 818 LQGGKGLFQSDPVLFSTSGAATVAIVNSFSSNPTLFFENFKASMIKMSRIGVLTGSQGEI 997
Query: 313 RKNCRVVN 320
RK C VN
Sbjct: 998 RKPCNFVN 1021
>TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase
precursor , partial (66%)
Length = 738
Score = 234 bits (598), Expect = 1e-62
Identities = 119/234 (50%), Positives = 155/234 (65%), Gaps = 1/234 (0%)
Frame = +1
Query: 4 SLSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
+LS L+ L+ +A L+ FY+ +CPK +R+A+ + KE RMGAS+LRL F
Sbjct: 52 ALSILLSLLAC----SANAQLTPTFYDRSCPKLQTIVRNAMVQTIKKEARMGASILRLFF 219
Query: 64 HDCFVQGCDASVLLDDTDS-FTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADI 122
HDCFV GCD S+LLDD + F GEKN+ PN NS RGFEVID IK +E+ C VSCADI
Sbjct: 220 HDCFVNGCDGSILLDDIGTTFVGEKNAAPNKNSARGFEVIDTIKTNVEASCNNTVSCADI 399
Query: 123 LTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNAT 182
L +A RD + LGG W + LGRRD+ TAS +N+++P+P DLS LIS + KG +A
Sbjct: 400 LALATRDGINLLGGPTWQVPLGRRDARTASQSKANTEIPSPSSDLSTLISMFSAKGLSAR 579
Query: 183 EMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASP 236
++ LSG HTIG + C F+R R+ NETNID +FAAS + NC GG D N +P
Sbjct: 580 DLTVLSGGHTIGQAECQFFRSRVNNETNIDAAFAASRKTNCPASGGG-DTNLAP 738
>TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (85%)
Length = 1352
Score = 230 bits (586), Expect = 3e-61
Identities = 127/301 (42%), Positives = 183/301 (60%), Gaps = 4/301 (1%)
Frame = +3
Query: 24 LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSF 83
LS FY+ +CPK IR ++ K+ A LLRLHFHDCFVQGCDASVLLD + S
Sbjct: 138 LSWTFYDSSCPKLESIIRKELKKVFDKDIAQAAGLLRLHFHDCFVQGCDASVLLDGSASG 317
Query: 84 TGEKNSFPNAN-SLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQ 142
EK++ PN + F++I+D++ Q+E C VVSCADI +AARD+VF GG ++L
Sbjct: 318 PSEKDAPPNLTLRAQAFKIIEDLRSQIEKKCGRVVSCADITAIAARDAVFLSGGPDYELP 497
Query: 143 LGRRDSTT-ASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFY 201
LGRRD A+ + + +LPAP + + ++++ K + T++V+LSG HTIG+S C +
Sbjct: 498 LGRRDGLNFATRNVTLDNLPAPQSNTTTILNSLATKNLDPTDVVSLSGGHTIGISHCSSF 677
Query: 202 RDRIY--NETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQKGL 259
DR+Y + +D +F ++++ C D N + D + FDN +Y +L+N++GL
Sbjct: 678 TDRLYPSKDPVMDQTFEKNLKLTCPASNTD---NTTVLDLRSPNTFDNKYYVDLMNRQGL 848
Query: 260 VHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVV 319
SDQ LY + T VTS++ N F E F AM+KM L+ LTGS G+IR NC V
Sbjct: 849 FFSDQDLYTD--KRTKDIVTSFAVNQSLFFEKFVVAMLKMGQLNVLTGSQGEIRANCSVR 1022
Query: 320 N 320
N
Sbjct: 1023N 1025
>TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%)
Length = 559
Score = 194 bits (492), Expect = 2e-50
Identities = 98/166 (59%), Positives = 124/166 (74%), Gaps = 2/166 (1%)
Frame = +3
Query: 8 LIFLITICLVGKT-SAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDC 66
++ L + L+G + SA LS FY + CP ++S V +AV+KE RMG SLLRL FHDC
Sbjct: 60 VLSLFMLFLIGSSNSAQLSENFYVKKCPSVFNAVKSVVHSAVAKEARMGGSLLRLFFHDC 239
Query: 67 FVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVA 126
FV GCD SVLLDDT SF GEK + PN+NSLRGF+VID IK ++E++CPGVVSCAD++ +A
Sbjct: 240 FVNGCDGSVLLDDTSSFKGEKTAPPNSNSLRGFDVIDAIKSKVEAVCPGVVSCADVVAIA 419
Query: 127 ARDSVFALGGQRWDLQLGRRDSTTASLDASNSD-LPAPFLDLSGLI 171
ARDSV LGG W ++LGRRDS TAS +A+NS +P+PF LS LI
Sbjct: 420 ARDSVAILGGPYWKVKLGRRDSKTASFNAANSGVIPSPFSSLSDLI 557
>TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precursor (Atperox
P10) (ATP5a) , partial (42%)
Length = 753
Score = 192 bits (489), Expect = 5e-50
Identities = 97/191 (50%), Positives = 128/191 (66%), Gaps = 1/191 (0%)
Frame = +3
Query: 9 IFLITICLVGK-TSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCF 67
+F++ + L+ S+ L FY+ TCP R +R + +A+S + R+ ASLLRLHFHDCF
Sbjct: 177 VFMLWLVLLSPLVSSQLYYNFYDTTCPNLTRIVRYNLLSAMSNDTRIAASLLRLHFHDCF 356
Query: 68 VQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAA 127
V GCD SVLLDDT + GEKN+ PN NS+RGFEVID IK LE CP VSCADILT+AA
Sbjct: 357 VNGCDGSVLLDDTSTQKGEKNALPNKNSIRGFEVIDTIKSALEKACPSTVSCADILTLAA 536
Query: 128 RDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTL 187
R++V+ G W + LGRRD TTAS +N +LP+PF L + + + KG ++ L
Sbjct: 537 REAVYLSRGPFWSVPLGRRDGTTASESEAN-NLPSPFEPLENITAKFISKGLEKKDVAVL 713
Query: 188 SGAHTIGLSRC 198
SGAHT G ++C
Sbjct: 714 SGAHTFGFAQC 746
>TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), partial (86%)
Length = 1286
Score = 191 bits (486), Expect = 1e-49
Identities = 109/299 (36%), Positives = 166/299 (55%), Gaps = 2/299 (0%)
Frame = +1
Query: 24 LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSF 83
LS FY++TCPK +R+ ++ + K+ LLR+ FHDCFVQGCD SVLLD +
Sbjct: 106 LSFSFYSKTCPKLETVVRNHLKKVLKKDNGQAPGLLRIFFHDCFVQGCDGSVLLDGS--- 276
Query: 84 TGEKNSFPNAN-SLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQ 142
GE++ N + I+DI+ + C +VSCADI +A+RD+VF GG + +
Sbjct: 277 PGERDQPANIGIRPEALQTIEDIRALVHKQCGKIVSCADITILASRDAVFLTGGPDYAVP 456
Query: 143 LGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYR 202
LGRRD + S LP+P + + + A+ + F+AT++V LSGAHT G + C +
Sbjct: 457 LGRRDGVSFS-TVGTQKLPSPINNTTATLKAFADRNFDATDVVALSGAHTFGRAHCGTFF 633
Query: 203 DRIYN-ETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQKGLVH 261
+R+ + N+D + A ++ C N + D T FDN +Y +L+N++G+
Sbjct: 634 NRLSPLDPNMDKTLAKNLTATCP---AQNSTNTANLDIRTPNVFDNKYYLDLMNRQGVFT 804
Query: 262 SDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
SDQ L T V +++ N F E F +A++K+S L LTG+ G+IR C VVN
Sbjct: 805 SDQDLL--SDKRTKGLVNAFAVNQTLFFEKFVDAVIKLSQLDVLTGNQGEIRGRCNVVN 975
>BP051104
Length = 566
Score = 188 bits (477), Expect(2) = 2e-49
Identities = 91/165 (55%), Positives = 116/165 (70%)
Frame = +2
Query: 68 VQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAA 127
V GCDAS+L DDT +F GE+ + N S RGF VID IK LE CPGVVSCAD+L +AA
Sbjct: 71 VNGCDASILXDDTSNFIGEQTAAANNRSARGFNVIDGIKANLEKQCPGVVSCADVLALAA 250
Query: 128 RDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTL 187
RDSV LGG W++ LGRRDSTTAS +N+ +P PFL LSGLI+ + +G + T++V L
Sbjct: 251 RDSVVQLGGPSWEVGLGRRDSTTASRGTANNTIPGPFLSLSGLITNFANQGLSVTDLVAL 430
Query: 188 SGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDN 232
SGAHTIGL++C +R IYN++NID S+A ++ C G D N
Sbjct: 431 SGAHTIGLAQCKNFRAHIYNDSNIDASYAKFLKX*CPRSGNDNLN 565
Score = 24.3 bits (51), Expect(2) = 2e-49
Identities = 11/17 (64%), Positives = 13/17 (75%)
Frame = +1
Query: 44 VEAAVSKEPRMGASLLR 60
V A+ KE R+GASLLR
Sbjct: 1 VAKAIQKEARIGASLLR 51
>TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%)
Length = 645
Score = 188 bits (477), Expect = 1e-48
Identities = 96/203 (47%), Positives = 131/203 (64%), Gaps = 1/203 (0%)
Frame = +2
Query: 6 SCLIFLITICLVGKTS-AVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFH 64
+C I ++ I L+ + + + L +Y+ +C A ++ V +V+K P + A L+R+HFH
Sbjct: 26 NCAIIVLVIFLLNQNAYSELEVGYYSYSCGMAEFIVKDEVRRSVTKNPGIAAGLVRMHFH 205
Query: 65 DCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILT 124
DCF++GCDASVLLD T S T EK+S N SLRGFEVID K +LE++C GVVSCADI+
Sbjct: 206 DCFIRGCDASVLLDSTPSNTAEKDSPANKPSLRGFEVIDSAKAKLEAVCKGVVSCADIIA 385
Query: 125 VAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEM 184
AARDSV GG +D+ GRRD + + +DLP P +++ L + KKG EM
Sbjct: 386 FAARDSVELAGGVGYDVPAGRRDGRISLASDTRTDLPPPTFNVNQLTQLFAKKGLTQEEM 565
Query: 185 VTLSGAHTIGLSRCIFYRDRIYN 207
VTLSGAHTIG S C + R+YN
Sbjct: 566 VTLSGAHTIGRSHCAAFSSRLYN 634
>TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (49%)
Length = 581
Score = 183 bits (464), Expect = 4e-47
Identities = 93/176 (52%), Positives = 131/176 (73%), Gaps = 5/176 (2%)
Frame = +2
Query: 8 LIFLITIC--LVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHD 65
++ L+T+ L+ T A L+ +Y++ CP+AL I+S V+ A+++E R+GASLLRLHFHD
Sbjct: 53 VLVLVTLATFLIPSTLAGLTPGYYDKVCPQALPIIKSVVQKAINREQRIGASLLRLHFHD 232
Query: 66 CFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCP-GVVSCADILT 124
CFV GCD S+LLDDT SF GEK + PN NS+RGFEV+D+IK ++ C VVSCADIL
Sbjct: 233 CFVNGCDGSLLLDDTSSFVGEKTALPNLNSVRGFEVVDEIKAAVDRACKRPVVSCADILA 412
Query: 125 VAARDSVFALGGQR--WDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKG 178
+AARDSV LGG++ + ++LGRRD+ +AS A+NS+LP PFL+ + L+ A+ +G
Sbjct: 413 IAARDSVAILGGKQYWYQVRLGRRDARSASQAAANSNLPPPFLNFTQLLPAFQAQG 580
>TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), partial
(68%)
Length = 614
Score = 182 bits (462), Expect = 7e-47
Identities = 98/193 (50%), Positives = 125/193 (63%), Gaps = 2/193 (1%)
Frame = +3
Query: 28 FYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSFTGEK 87
FY TCP+A +RS VE+ V+ +P + A LLR+HFHDCFVQGCDASVL+ + E+
Sbjct: 36 FYLGTCPRAESIVRSTVESHVNSDPTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 206
Query: 88 NSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQLGRRD 147
+ PN SLRGFEVIDD K ++E+ CPGVVSCADIL +AARDSV GG W + GRRD
Sbjct: 207 TAIPNL-SLRGFEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 383
Query: 148 STTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRDRIYN 207
+ N +LPAPF + + KG N ++VTL G HTIG + C F+ +R+YN
Sbjct: 384 GRVSQASDVN-NLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 560
Query: 208 ETN--IDPSFAAS 218
T+ DPS AS
Sbjct: 561 FTSNGPDPSIDAS 599
>BG662240
Length = 312
Score = 179 bits (455), Expect = 4e-46
Identities = 89/103 (86%), Positives = 94/103 (90%)
Frame = +3
Query: 107 KQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLD 166
KQLESMCPGVVSCADILT+AARDSV ALGG+RW+L LGRRDSTTASLDASNSDLPAPFLD
Sbjct: 3 KQLESMCPGVVSCADILTIAARDSVVALGGERWNLLLGRRDSTTASLDASNSDLPAPFLD 182
Query: 167 LSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRDRIYNET 209
LSGLISA+ KKGF EMVTLSGAHTIGL RC+F R RIYNET
Sbjct: 183 LSGLISAFDKKGFTTAEMVTLSGAHTIGLVRCLFTRARIYNET 311
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 179 bits (455), Expect = 4e-46
Identities = 111/302 (36%), Positives = 156/302 (50%), Gaps = 5/302 (1%)
Frame = +2
Query: 24 LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSF 83
L+ FY ETCP+A I V+ + S LR FHDC VQ CDAS+LLD T
Sbjct: 152 LAMNFYKETCPQAEDIITEQVKLLYKRHKNTAFSWLRNIFHDCAVQSCDASLLLDSTRKT 331
Query: 84 TGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQL 143
EK + + LR F I+ IK+ LE CPGVVSC+DIL ++ARD + ALGG L+
Sbjct: 332 LSEKET-DRSFGLRNFRYIETIKEALERECPGVVSCSDILVLSARDGIAALGGPHIPLRT 508
Query: 144 GRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRD 203
GRRD + D LP +S ++ +G G + +V L GAH++G + C+
Sbjct: 509 GRRDGRRSRADVVEQFLPDHNESISAVLDKFGAMGIDTPGVVALLGAHSVGRTHCVKLVH 688
Query: 204 RIYNETN--IDPSFAASMQVNCSFDGGDTDNNASPF---DSTTQFKFDNAFYQNLLNQKG 258
R+Y E + ++P M C D A + D T DN +Y+N+L+ KG
Sbjct: 689 RLYPEVDPALNPDHIPHMLKKC--PDAIPDPKAVQYVRNDRGTPMILDNNYYRNILDNKG 862
Query: 259 LVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRV 318
L+ D QL T V +K+ F ++F+ A+ +S +PLTG+ G+IRK C V
Sbjct: 863 LLLVDHQL--ANDKRTKPYVKKMAKSQDYFFKEFSRAITLLSENNPLTGTKGEIRKQCNV 1036
Query: 319 VN 320
N
Sbjct: 1037AN 1042
>TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , partial
(96%)
Length = 1390
Score = 172 bits (437), Expect = 5e-44
Identities = 113/321 (35%), Positives = 169/321 (52%), Gaps = 13/321 (4%)
Frame = +1
Query: 12 ITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGC 71
+T+C A LS Y TCP ++ V+ + + LRL FHDCFVQGC
Sbjct: 94 LTLCFY-TCFAQLSPNHYASTCPNLQSIVKGVVQKKFQQTFVTVPATLRLFFHDCFVQGC 270
Query: 72 DASVLLDDTDSFTGEKNSFPNANSLRG--FEVIDDIKKQLESM--CPGVVSCADILTVAA 127
DASV++ + + EK+ P+ SL G F+ + K ++++ C VSCADIL +A
Sbjct: 271 DASVMVASSGNNKAEKDH-PDNLSLAGDGFDTVIKAKAAVDAVPQCRNKVSCADILALAT 447
Query: 128 RDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTL 187
RD V GG + ++LGR D + N LP P +L+ L S + +G T+M+ L
Sbjct: 448 RDVVVLAGGPSYTVELGRFDGLVSRASDVNGRLPEPNFNLNQLNSLFASQGLTQTDMIAL 627
Query: 188 SGAHTIGLSRCIFYRDRIYNETNIDP----SFAASMQVNCSFDGGDTDNNASP-----FD 238
SGAHT+G S C + +RIY+ T +DP ++A +Q C N +P D
Sbjct: 628 SGAHTLGFSHCNRFSNRIYS-TPVDPTLNRNYATQLQQMC-------PKNVNPQIAINMD 783
Query: 239 STTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVK 298
TT FDN +Y+NL KGL SDQ L+ + + + V S++ N+ F +FA AM+K
Sbjct: 784 PTTPRTFDNIYYKNLQQGKGLFTSDQILFTD--QRSKATVNSFASNSNTFNANFAAAMIK 957
Query: 299 MSLLSPLTGSDGQIRKNCRVV 319
+ + T +G+IR +C V+
Sbjct: 958 LGRVGVKTARNGKIRTDCSVL 1020
>TC18696 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (47%)
Length = 500
Score = 169 bits (429), Expect = 4e-43
Identities = 87/153 (56%), Positives = 112/153 (72%), Gaps = 3/153 (1%)
Frame = +2
Query: 8 LIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCF 67
++ ++T+ LV + A LS FYN+ CP+AL I S V A+ +E R+GASLLRLHFHDCF
Sbjct: 41 VLVMVTLTLVIPSKAQLSPSFYNKVCPQALPVINSVVRRAILRERRIGASLLRLHFHDCF 220
Query: 68 VQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCP-GVVSCADILTVA 126
V GCD SVLLDDT +F GEK +FPN NS++GF+V+D+IKK ++ C VVSCADIL +A
Sbjct: 221 VNGCDGSVLLDDTRNFIGEKTAFPNNNSIKGFDVVDEIKKAVDKACKRPVVSCADILAIA 400
Query: 127 ARDSVFALGGQR--WDLQLGRRDSTTASLDASN 157
ARDSV LGG + + LGRRD+ TAS A+N
Sbjct: 401 ARDSVAILGGPSLVYKVLLGRRDARTASRAAAN 499
>TC10510 similar to PIR|B56555|B56555 peroxidase , anionic, precursor - wood
tobacco {Nicotiana sylvestris;} , partial (53%)
Length = 764
Score = 155 bits (393), Expect = 7e-39
Identities = 87/187 (46%), Positives = 117/187 (62%), Gaps = 2/187 (1%)
Frame = +3
Query: 136 GQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGL 195
G W ++LGRRDSTTAS +N++LP+ DL LIS + KG A +MV LSGAHTIG
Sbjct: 3 GPSWTVKLGRRDSTTASRSLANTELPSFTDDLQTLISRFDNKGLTARDMVILSGAHTIGQ 182
Query: 196 SRCIFYRDRIYNE-TNIDPSFAASMQVNC-SFDGGDTDNNASPFDSTTQFKFDNAFYQNL 253
++C +R RIYN ++ID F ++ + C S + D + D T FDN +++NL
Sbjct: 183 AQCSTFRGRIYNNASDIDAGFGSTRRRGCPSTISVENDKKLAALDLVTPNSFDNNYFKNL 362
Query: 254 LNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIR 313
+ +KGL+ SDQ L+ GS TD+ V+ YSKN F DFA AM+KM + PLTGS G IR
Sbjct: 363 IQKKGLLRSDQVLFRGGS--TDTIVSEYSKNPTTFKSDFAAAMIKMGDIQPLTGSAGIIR 536
Query: 314 KNCRVVN 320
K C +N
Sbjct: 537 KICSAIN 557
>TC9437 similar to UP|Q7XYR7 (Q7XYR7) Class III peroxidase, partial (40%)
Length = 589
Score = 150 bits (378), Expect = 4e-37
Identities = 79/137 (57%), Positives = 96/137 (69%), Gaps = 1/137 (0%)
Frame = +1
Query: 185 VTLSGAHTIGLSRCIFYRDRIYNETN-IDPSFAASMQVNCSFDGGDTDNNASPFDSTTQF 243
V LSGAHTIG +RC +R RIYNET+ I+ SFA S + NC G DNN +P D T
Sbjct: 1 VALSGAHTIGQARCTSFRARIYNETSTIESSFATSRKSNCPSTSGSGDNNLAPLDLQTPT 180
Query: 244 KFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLS 303
FDN +++NL+ KGL+HSDQQL+ NG G+TDS V YS N F DFA+AMVKM +S
Sbjct: 181 SFDNNYFKNLVQNKGLLHSDQQLF-NG-GSTDSTVRGYSTNPSSFSSDFASAMVKMGDIS 354
Query: 304 PLTGSDGQIRKNCRVVN 320
PLTGS+G+IRKNCR N
Sbjct: 355 PLTGSNGEIRKNCRKTN 405
>TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
partial (47%)
Length = 604
Score = 144 bits (362), Expect = 3e-35
Identities = 75/163 (46%), Positives = 104/163 (63%), Gaps = 2/163 (1%)
Frame = +1
Query: 3 GSLSCLIFLIT--ICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLR 60
G SC LI + L+ A L FY +TCPKA + I V + P + A+L+R
Sbjct: 94 GGQSCFKVLIVCLLALIASNHAQLEQGFYTKTCPKAEKIILDFVHEHIHNAPSLAAALIR 273
Query: 61 LHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCA 120
++FHDCFV+GCDAS+LL+ T S E+++ PN ++RGF+ ID IK +E+ CPGVVSCA
Sbjct: 274 MNFHDCFVRGCDASILLNST-SKQAERDAPPNL-TVRGFDFIDRIKSLVEAQCPGVVSCA 447
Query: 121 DILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAP 163
DI+ +AARDS+ A GG W + GRRD ++L + S +PAP
Sbjct: 448 DIIALAARDSIVATGGPFWKVPTGRRDGVISNLVEARSQIPAP 576
>TC17568 similar to UP|Q8RVP6 (Q8RVP6) Gaiacol peroxidase , partial (33%)
Length = 672
Score = 134 bits (337), Expect = 2e-32
Identities = 65/124 (52%), Positives = 88/124 (70%)
Frame = +2
Query: 8 LIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCF 67
LIFL+ + +S+ L FY+ TCP+A ++ ++ A+ +EPR AS++R FHDCF
Sbjct: 296 LIFLLHTTSLVTSSSDLRPGFYSNTCPEAEYIVQDVMKKALFREPRSVASVMRFQFHDCF 475
Query: 68 VQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAA 127
V GCDAS+LLDDT GEK + N NSLR FEV+D+IK+ LE CPGVVSCADI+ +A+
Sbjct: 476 VNGCDASMLLDDTPDMLGEKLALSNINSLRSFEVVDEIKEALEKKCPGVVSCADIIIMAS 655
Query: 128 RDSV 131
RD+V
Sbjct: 656 RDAV 667
>TC18385 similar to UP|Q9XFL6 (Q9XFL6) Peroxidase 5, partial (38%)
Length = 424
Score = 134 bits (336), Expect = 3e-32
Identities = 67/130 (51%), Positives = 90/130 (68%), Gaps = 1/130 (0%)
Frame = +3
Query: 8 LIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCF 67
L+ L+TI ++ L++ FY+ TCP + +AV+ A+ + R+GASL+RLHFHDCF
Sbjct: 39 LVLLLTILF--PSNGQLNSTFYSSTCPNVSFIVSNAVQQALQSDSRIGASLIRLHFHDCF 212
Query: 68 VQGCDASVLLDDTDSFT-GEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVA 126
V G DAS+LLD + T EKN+ PN NS+RGF+V+D IK LE+ CPG+VSCADIL +A
Sbjct: 213 VNGGDASILLDQGGNITQSEKNAAPNFNSIRGFDVVDTIKSALENSCPGIVSCADILALA 392
Query: 127 ARDSVFALGG 136
A SV GG
Sbjct: 393 AESSVSMSGG 422
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,102,785
Number of Sequences: 28460
Number of extensions: 62064
Number of successful extensions: 408
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 347
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 349
length of query: 320
length of database: 4,897,600
effective HSP length: 90
effective length of query: 230
effective length of database: 2,336,200
effective search space: 537326000
effective search space used: 537326000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0260.7


