

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.11
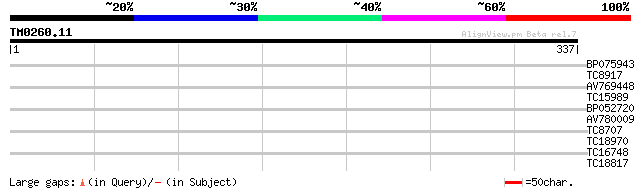
(337 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP075943 35 0.018
TC8917 similar to UP|AAH64262 (AAH64262) MGC76273 protein, parti... 32 0.20
AV769448 29 0.98
TC15989 28 2.2
BP052720 28 2.9
AV780009 27 3.7
TC8707 weakly similar to UP|Q9FN67 (Q9FN67) Gb|AAD56319.1, parti... 27 3.7
TC18970 weakly similar to UP|Q8I8F0 (Q8I8F0) Chromosome scaffold... 27 4.9
TC16748 similar to GB|AAO64917.1|29029076|BT005982 At4g28770 {Ar... 27 6.4
TC18817 26 8.3
>BP075943
Length = 547
Score = 35.0 bits (79), Expect = 0.018
Identities = 37/121 (30%), Positives = 51/121 (41%), Gaps = 25/121 (20%)
Frame = -3
Query: 138 LHSVFPIKDLGELHYFL-MKHCKSASTLMIHDFVCQMSVLFNLT*HFVKKRSILALLSIQ 196
LH F IKDLGE YFL ++ +S S ++++ +++ L L+S
Sbjct: 449 LHDQFRIKDLGEAKYFLGLEIARSTSGIVLN-----------------QRKYALQLISDS 321
Query: 197 *NSFGFS--FNSH----------------DSWLYKSIVGAL------HPDIAFCVNKVCQ 232
FGF F SH D Y+ IVG L PDI F VN++ Q
Sbjct: 320 -GHFGFQPRFYSHGTNSQTLGTNTGTPLTDIGSYRRIVGRLLYLNTTRPDITFAVNQLSQ 144
Query: 233 F 233
F
Sbjct: 143 F 141
>TC8917 similar to UP|AAH64262 (AAH64262) MGC76273 protein, partial (3%)
Length = 676
Score = 31.6 bits (70), Expect = 0.20
Identities = 26/75 (34%), Positives = 41/75 (54%)
Frame = -1
Query: 105 IGLLPV*LLFLCFMLGYNNPPLFFLTHLFFYLGLHSVFPIKDLGELHYFLMKHCKSASTL 164
+GLL L F + LG+N L+FL+ FF+LGL F ++ L L + L++ +
Sbjct: 424 LGLL---LGFRVYFLGFN---LYFLSGCFFFLGLSLYFFLRFLC-LLFILIEQGA*RLVI 266
Query: 165 MIHDFVCQMSVLFNL 179
I F+C +LFN+
Sbjct: 265 FIRGFLC---LLFNI 230
>AV769448
Length = 447
Score = 29.3 bits (64), Expect = 0.98
Identities = 16/38 (42%), Positives = 21/38 (55%)
Frame = +1
Query: 128 FLTHLFFYLGLHSVFPIKDLGELHYFLMKHCKSASTLM 165
F +HL FY LHS PI L L+ H KS ++L+
Sbjct: 37 FFSHLPFYPPLHSPSPISSSPRLTVVLI*HHKSRASLL 150
>TC15989
Length = 526
Score = 28.1 bits (61), Expect = 2.2
Identities = 13/43 (30%), Positives = 22/43 (50%)
Frame = +2
Query: 132 LFFYLGLHSVFPIKDLGELHYFLMKHCKSASTLMIHDFVCQMS 174
+FFY+ P+K +H MKHCK+ + + + C+ S
Sbjct: 236 IFFYI------PVKVFFHVHILGMKHCKANALFCVQE*FCRHS 346
>BP052720
Length = 510
Score = 27.7 bits (60), Expect = 2.9
Identities = 13/30 (43%), Positives = 20/30 (66%)
Frame = +1
Query: 189 ILALLSIQ*NSFGFSFNSHDSWLYKSIVGA 218
++ LLS+ NS+ F F+S SW ++VGA
Sbjct: 274 LMLLLSLLFNSWTFYFSSIPSWATTNVVGA 363
>AV780009
Length = 529
Score = 27.3 bits (59), Expect = 3.7
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = -1
Query: 236 LIFLTAKKVVVVS*SNTEAEYRSIAFDTI*PIWLQF 271
LI KK VS S++EAEYR++A WL +
Sbjct: 361 LISWRTKKQTTVSRSSSEAEYRALAATVCEVQWLSY 254
>TC8707 weakly similar to UP|Q9FN67 (Q9FN67) Gb|AAD56319.1, partial (43%)
Length = 1062
Score = 27.3 bits (59), Expect = 3.7
Identities = 13/38 (34%), Positives = 20/38 (52%)
Frame = -1
Query: 126 LFFLTHLFFYLGLHSVFPIKDLGELHYFLMKHCKSAST 163
++ + H F Y LHS FP + L L+ L+ C+ T
Sbjct: 774 MYKVLHNFIYKKLHSKFPRRTLPNLNTLLILPCQEHPT 661
>TC18970 weakly similar to UP|Q8I8F0 (Q8I8F0) Chromosome scaffold protein
p85, partial (4%)
Length = 562
Score = 26.9 bits (58), Expect = 4.9
Identities = 17/43 (39%), Positives = 23/43 (52%)
Frame = -3
Query: 112 LLFLCFMLGYNNPPLFFLTHLFFYLGLHSVFPIKDLGELHYFL 154
L F F+LG+ FFL F+L LH F + D LH++L
Sbjct: 233 LFFTLFLLGF-----FFLLLHHFFLVLHHFFLVLDF--LHFWL 126
>TC16748 similar to GB|AAO64917.1|29029076|BT005982 At4g28770 {Arabidopsis
thaliana;}, partial (36%)
Length = 442
Score = 26.6 bits (57), Expect = 6.4
Identities = 16/44 (36%), Positives = 20/44 (45%)
Frame = +3
Query: 115 LCFMLGYNNPPLFFLTHLFFYLGLHSVFPIKDLGELHYFLMKHC 158
LC ++ PP L HL F+ D+ LHYFL HC
Sbjct: 237 LCHQCQFSQPPCS-LVHLCFH-------GCGDIDVLHYFLGLHC 344
>TC18817
Length = 770
Score = 26.2 bits (56), Expect = 8.3
Identities = 10/28 (35%), Positives = 17/28 (60%)
Frame = +1
Query: 113 LFLCFMLGYNNPPLFFLTHLFFYLGLHS 140
L LC++L Y+ + +TH+ FY H+
Sbjct: 88 LRLCWVLSYSFNSILIITHVLFYKRKHN 171
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.364 0.164 0.596
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,672,404
Number of Sequences: 28460
Number of extensions: 105664
Number of successful extensions: 1634
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1614
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1633
length of query: 337
length of database: 4,897,600
effective HSP length: 91
effective length of query: 246
effective length of database: 2,307,740
effective search space: 567704040
effective search space used: 567704040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0260.11


