

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
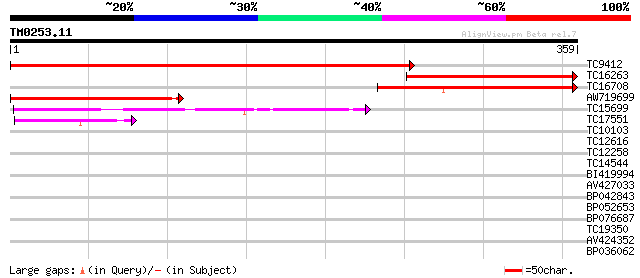
Query= TM0253.11
(359 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9412 weakly similar to GB|AAL15368.1|16323268|AY057738 At1g156... 535 e-153
TC16263 weakly similar to UP|Q7PYA9 (Q7PYA9) AgCP12622 (Fragment... 233 5e-62
TC16708 154 3e-38
AW719699 141 2e-34
TC15699 similar to UP|O64797 (O64797) T1F15.5 protein, partial (... 77 4e-15
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 42 2e-04
TC10103 weakly similar to AAS47616 (AAS47616) At4g39570, partial... 32 0.13
TC12616 32 0.22
TC12258 weakly similar to UP|O50458 (O50458) PGRS_FAMILY protein... 30 0.82
TC14544 30 0.82
BI419994 29 1.1
AV427033 28 1.8
BP042843 28 2.4
BP052653 28 2.4
BP076687 28 3.1
TC19350 similar to UP|Q9MAL7 (Q9MAL7) T25K16.17, partial (20%) 27 4.1
AV424352 27 6.9
BP036062 26 9.0
>TC9412 weakly similar to GB|AAL15368.1|16323268|AY057738 At1g15670/F7H2_1
{Arabidopsis thaliana;}, partial (30%)
Length = 961
Score = 535 bits (1379), Expect = e-153
Identities = 255/256 (99%), Positives = 255/256 (99%)
Frame = +1
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELISGLPEDV RDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA
Sbjct: 193 MELISGLPEDVPRDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 372
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 120
QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY
Sbjct: 373 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 552
Query: 121 ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 180
ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD
Sbjct: 553 ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 732
Query: 181 EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 240
EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS
Sbjct: 733 EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 912
Query: 241 AEEFDVDTWQWGPVEE 256
AEEFDVDTWQWGPVEE
Sbjct: 913 AEEFDVDTWQWGPVEE 960
>TC16263 weakly similar to UP|Q7PYA9 (Q7PYA9) AgCP12622 (Fragment), partial
(23%)
Length = 605
Score = 233 bits (593), Expect = 5e-62
Identities = 108/108 (100%), Positives = 108/108 (100%)
Frame = +2
Query: 252 GPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVAKVPSEIRNVACMGAWE 311
GPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVAKVPSEIRNVACMGAWE
Sbjct: 14 GPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVAKVPSEIRNVACMGAWE 193
Query: 312 RSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLLEI 359
RSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLLEI
Sbjct: 194 RSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLLEI 337
>TC16708
Length = 515
Score = 154 bits (388), Expect = 3e-38
Identities = 76/130 (58%), Positives = 96/130 (73%), Gaps = 4/130 (3%)
Frame = +3
Query: 234 QGRFERSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGD---AMYMCRGG-DVVALQGN 289
QG FER+AE FD +WG VEE FL C TCPRT +DGGD + YMC GG D++A +G+
Sbjct: 3 QGGFERTAEAFDAAEGKWGQVEENFLGCATCPRTVVDGGDGDESGYMCSGGGDLMAKRGD 182
Query: 290 TWQTVAKVPSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTG 349
TWQ VA+VP EI NVA +GA + S+LLIGS+G+G HMG++ D+KS W KL SP+ + G
Sbjct: 183 TWQKVARVPGEICNVAYVGALDGSVLLIGSNGYGGVHMGYVFDVKSCSWRKLESPEGFHG 362
Query: 350 HVQSYCLLEI 359
HVQS C+LEI
Sbjct: 363 HVQSGCILEI 392
>AW719699
Length = 489
Score = 141 bits (355), Expect = 2e-34
Identities = 70/110 (63%), Positives = 82/110 (73%)
Frame = +1
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELISGLP+DVARDCLIRVSYEQF A A C+GW TEI SPEF R+RR TG+ +K++VT
Sbjct: 160 MELISGLPDDVARDCLIRVSYEQFAAAAAVCRGWKTEIHSPEFHRQRRSTGHAEKVMVTV 339
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLP 110
QA+ + + G K TNPVYRLS+F TG+WSEL PP F GSGLP
Sbjct: 340 QAQPEPEKSGTGSTKRLTNPVYRLSLFNLGTGDWSELPAPPGF--GSGLP 483
>TC15699 similar to UP|O64797 (O64797) T1F15.5 protein, partial (53%)
Length = 902
Score = 77.0 bits (188), Expect = 4e-15
Identities = 67/229 (29%), Positives = 102/229 (44%), Gaps = 3/229 (1%)
Frame = +1
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
++ GLP+DV++ CL V FP + CK W IQS EF R++ G ++ L
Sbjct: 148 ILPGLPDDVSKHCLALVPRSNFPTMGGVCKKWRRFIQSKEFLTVRKLAGQVEEWLY---- 315
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELT-MPPEFDSGSGLPMFCQIAGVGYE 121
+L+ + V + N L MP +G G+ + + +
Sbjct: 316 ---------ILIAGSEGKGSHWEVMDCLGHNRRSLPPMPGPDKTGFGVVV------LNGK 450
Query: 122 LVVMGGWDPESWKASNSVFIYNFLSA--TWRRGADMPGGPRTFFACTSDQDRTVYVAGGH 179
L+VM G+ S S +Y + S +W R +M R FAC ++ + VY GG+
Sbjct: 451 LLVMAGYASIDGIVSVSAEVYQYNSCLNSWTRLPNM-NVARYDFAC-AEVNGLVYAVGGY 624
Query: 180 DEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGG 228
E ++L SA YD D+W + + R R C A G GKL V+GG
Sbjct: 625 GEHGDSLSSAEVYDPDTDKWTLIESLRRPRWGCFAC--GIEGKLYVMGG 765
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 42.0 bits (97), Expect = 2e-04
Identities = 26/80 (32%), Positives = 39/80 (48%), Gaps = 3/80 (3%)
Frame = +2
Query: 4 ISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEF---RRRRRVTGNTQKILVTA 60
IS LP+D+ D L RV P+++ C+ W+ + SP+F RRRR + ++ L
Sbjct: 128 ISSLPDDIILDFLTRVPPSSLPSLSLVCRRWSHLLHSPDFSDLRRRRHLLRHSALALTAT 307
Query: 61 QARFDSDTCGGLLVKATTNP 80
S T L+ A NP
Sbjct: 308 DFGLSSAT----LLDAKWNP 355
>TC10103 weakly similar to AAS47616 (AAS47616) At4g39570, partial (15%)
Length = 318
Score = 32.3 bits (72), Expect = 0.13
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQ 54
LI LP+DVA +CL RV P ++ K + + SP F R + TQ
Sbjct: 93 LIPNLPDDVAVNCLARVPRSDHPNLSLVSKPIHFLLTSPLFFNARSLLNCTQ 248
>TC12616
Length = 434
Score = 31.6 bits (70), Expect = 0.22
Identities = 17/53 (32%), Positives = 26/53 (48%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQK 55
L+ GLP+D+A CLIRV + CK W + F R+ G +++
Sbjct: 228 LLPGLPDDLAIACLIRVPRVDHRKLRLVCKRWYRLLVGNFFYSLRKSLGISEE 386
>TC12258 weakly similar to UP|O50458 (O50458) PGRS_FAMILY protein (PE_PGRS
family protein), partial (6%)
Length = 471
Score = 29.6 bits (65), Expect = 0.82
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 5/51 (9%)
Frame = +1
Query: 38 IQSPEFRRRR-----RVTGNTQKILVTAQARFDSDTCGGLLVKATTNPVYR 83
+++P RRRR R T N+ + L+TA RF + T LLV A+T +R
Sbjct: 97 LRAPSPRRRRWHSSERSTANSPERLITA--RFSAATSNALLVLASTASAFR 243
>TC14544
Length = 1160
Score = 29.6 bits (65), Expect = 0.82
Identities = 32/131 (24%), Positives = 52/131 (39%), Gaps = 7/131 (5%)
Frame = +2
Query: 172 TVYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCT 231
T+Y+ GG + L D W+P P M R+ A G K+ V+GG
Sbjct: 8 TIYILGGSINDVPTPHVWLL-DCRFHRWLPGPSMRVAREFAAAGVIDG--KIYVIGGCIA 178
Query: 232 EMQGRFERSAEEFDVDTWQW------GPVEEEFLDCGTCPRTCLDGGDAMYMC-RGGDVV 284
+ R AE D T +W ++E+++ + + GG M RGG
Sbjct: 179 DNWTRSAHWAETLDPATGRWERVPSPPEIQEKWMHA-----SAVVGGKVYAMADRGGVAF 343
Query: 285 ALQGNTWQTVA 295
+ W++V+
Sbjct: 344 DPRSGAWESVS 376
Score = 29.3 bits (64), Expect = 1.1
Identities = 17/65 (26%), Positives = 25/65 (38%), Gaps = 2/65 (3%)
Frame = +2
Query: 70 GGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYELVVMGG-- 127
GG + T + +P TG W + PPE + + G Y + GG
Sbjct: 164 GGCIADNWTRSAHWAETLDPATGRWERVPSPPEIQE-KWMHASAVVGGKVYAMADRGGVA 340
Query: 128 WDPES 132
+DP S
Sbjct: 341 FDPRS 355
>BI419994
Length = 549
Score = 29.3 bits (64), Expect = 1.1
Identities = 12/45 (26%), Positives = 20/45 (43%)
Frame = +2
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRR 45
+ L S LP+++ L+R+ CK W I P+F +
Sbjct: 29 LSLFSHLPDELILQILLRLPVRSLLRFKTVCKSWRYLISDPQFTK 163
>AV427033
Length = 420
Score = 28.5 bits (62), Expect = 1.8
Identities = 12/39 (30%), Positives = 18/39 (45%)
Frame = +1
Query: 7 LPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRR 45
LP D+ + L R+ CK WN+ I P+F +
Sbjct: 124 LPFDLVVEILCRLPVNSLLQFRCVCKSWNSLISDPKFAK 240
>BP042843
Length = 452
Score = 28.1 bits (61), Expect = 2.4
Identities = 15/34 (44%), Positives = 17/34 (49%)
Frame = -3
Query: 44 RRRRRVTGNTQKILVTAQARFDSDTCGGLLVKAT 77
RRRRR+T IL+ R S CG KAT
Sbjct: 237 RRRRRITSGFGVILILTHRRRGSRKCGPAAKKAT 136
>BP052653
Length = 552
Score = 28.1 bits (61), Expect = 2.4
Identities = 13/43 (30%), Positives = 20/43 (46%)
Frame = +1
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEF 43
+ L S LP D+ L+R+ + CK W + I P+F
Sbjct: 169 LNLSSILPHDLIVQILLRLPVRSLLRFKSVCKSWLSLISDPQF 297
>BP076687
Length = 389
Score = 27.7 bits (60), Expect = 3.1
Identities = 12/39 (30%), Positives = 19/39 (47%)
Frame = -3
Query: 5 SGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEF 43
S LPE++ L+R+ + CK W + I P+F
Sbjct: 387 SHLPEELIPQILLRLPVRSLLRFKSVCKSWLSLISDPKF 271
>TC19350 similar to UP|Q9MAL7 (Q9MAL7) T25K16.17, partial (20%)
Length = 394
Score = 27.3 bits (59), Expect = 4.1
Identities = 15/33 (45%), Positives = 20/33 (60%)
Frame = +2
Query: 91 TGNWSELTMPPEFDSGSGLPMFCQIAGVGYELV 123
T N +E +P F SGSGL C++ GV +LV
Sbjct: 17 TRNLTESVLPVPFSSGSGLIWLCEL-GVFADLV 112
>AV424352
Length = 432
Score = 26.6 bits (57), Expect = 6.9
Identities = 9/22 (40%), Positives = 11/22 (49%)
Frame = +2
Query: 247 DTWQWGPVEEEFLDCGTCPRTC 268
DTW W ++ D GTC C
Sbjct: 239 DTWHWDVCKDVGSDSGTCSNLC 304
>BP036062
Length = 512
Score = 26.2 bits (56), Expect = 9.0
Identities = 15/55 (27%), Positives = 28/55 (50%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKIL 57
L+ GL +DVA +CL VS + +++ K ++ I S R+ G + ++
Sbjct: 321 LLPGLIDDVALNCLAWVSGSDYASLSCVNKRYSQLINSGYLYGLRKQVGAVEHLV 485
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,202,390
Number of Sequences: 28460
Number of extensions: 108750
Number of successful extensions: 434
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 430
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 433
length of query: 359
length of database: 4,897,600
effective HSP length: 91
effective length of query: 268
effective length of database: 2,307,740
effective search space: 618474320
effective search space used: 618474320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0253.11


