

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252d.2
(519 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
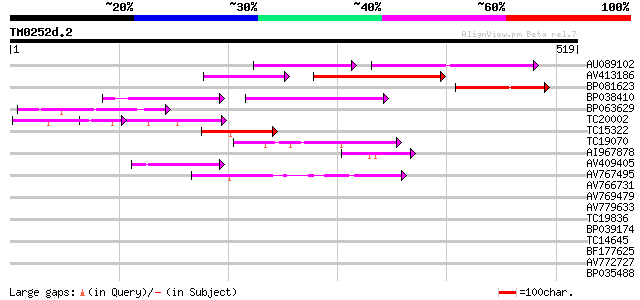
Score E
Sequences producing significant alignments: (bits) Value
AU089102 110 6e-25
AV413186 102 2e-22
BP081623 77 9e-15
BP038410 71 5e-13
BP063629 64 5e-11
TC20002 similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-l... 45 2e-05
TC15322 weakly similar to UP|Q9LG23 (Q9LG23) F14J16.14 (At1g5589... 45 4e-05
TC19070 weakly similar to UP|Q9LRP6 (Q9LRP6) DNA-binding protein... 43 1e-04
AI967878 43 1e-04
AV409405 42 2e-04
AV767495 41 5e-04
AV766731 39 0.003
AV769479 38 0.005
AV779633 37 0.010
TC19836 35 0.030
BP039174 31 0.43
TC14645 similar to UP|Q41122 (Q41122) Proline-rich protein precu... 30 0.73
BF177625 30 0.73
AV772727 30 1.2
BP035488 29 1.6
>AU089102
Length = 480
Score = 110 bits (275), Expect = 6e-25
Identities = 55/153 (35%), Positives = 88/153 (56%)
Frame = +2
Query: 332 GTQPTETTFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKE 391
G Q + T+V +LTAC+H G+V +G++ F+++ ++ + HYAC+VDL RAG +KE
Sbjct: 26 GFQANDVTYVELLTACSHAGLVDEGIQYFDKLLKNRSIQVKEXHYACLVDLCGRAGRLKE 205
Query: 392 AEKFMEKMGGLVEADLNVWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMY 451
+E +G V+ L+VWG +L C ++ N D+G V K++ + GT+ L NMY
Sbjct: 206 XFYIIEGLG--VKLSLSVWGPLLAGCNVHGNADIGKLVTTKILKVEHENAGTYSLLSNMY 379
Query: 452 REAGWDAEAKRVRSMVSEAGMKKKPGCSIIEVG 484
V +++ G+KK+P C IEVG
Sbjct: 380 ASXRXWKXXAPVXMKMNDKGLKKQPRCXWIEVG 478
Score = 43.1 bits (100), Expect = 1e-04
Identities = 24/96 (25%), Positives = 43/96 (44%), Gaps = 2/96 (2%)
Frame = +2
Query: 224 FEEMQNRGMKPNDSVLVTVLTACAHLGALTQGL-WVHSYAKRCKVDSNPILATALVDMYS 282
F Q G + ND V +LTAC+H G + +G+ + K + LVD+
Sbjct: 5 FGTRQELGFQANDVTYVELLTACSHAGLVDEGIQYFDKLLKNRSIQVKEXHYACLVDLCG 184
Query: 283 KCGCVESALTVFEGIADK-DAGAWNAMISGVALSGD 317
+ G ++ + EG+ K W +++G + G+
Sbjct: 185 RAGRLKEXFYIIEGLGVKLSLSVWGPLLAGCNVHGN 292
>AV413186
Length = 398
Score = 102 bits (254), Expect = 2e-22
Identities = 52/122 (42%), Positives = 75/122 (60%), Gaps = 1/122 (0%)
Frame = +1
Query: 279 DMYSKCGCVESALTVFEGIADK-DAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTE 337
DMYSKCG +E+A VF + +K D WN MIS +A G +++ + + M G +P
Sbjct: 4 DMYSKCGSLETAWRVFNLVGNKQDVVLWNTMISALAHHGFGTEAMVMLNDMIRSGVEPNR 183
Query: 338 TTFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFME 397
TFV +L AC+ G+V +GL+ F+ M+ +GV P EHYAC+ DLL RAG E+ K ++
Sbjct: 184 ATFVVLLNACSLSGLVQEGLQFFKFMTSEFGVVPDQEHYACLTDLLVRAGYFDESVKGLQ 363
Query: 398 KM 399
M
Sbjct: 364 IM 369
Score = 43.5 bits (101), Expect = 8e-05
Identities = 22/80 (27%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Frame = +1
Query: 178 DGYGKVGDVESARELFEEMPER-NAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPND 236
D Y K G +E+A +F + + + V W+ M++A + + + + +M G++PN
Sbjct: 4 DMYSKCGSLETAWRVFNLVGNKQDVVLWNTMISALAHHGFGTEAMVMLNDMIRSGVEPNR 183
Query: 237 SVLVTVLTACAHLGALTQGL 256
+ V +L AC+ G + +GL
Sbjct: 184 ATFVVLLNACSLSGLVQEGL 243
>BP081623
Length = 461
Score = 76.6 bits (187), Expect = 9e-15
Identities = 38/87 (43%), Positives = 54/87 (61%), Gaps = 1/87 (1%)
Frame = -2
Query: 409 VWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYRE-AGWDAEAKRVRSMV 467
VWGA+L ACRIY +V +G + + + CG +IL NMY E WD +A +VR ++
Sbjct: 460 VWGALLGACRIYGDVGLGRWAAEHLFELKPQHCGYYILLANMYAEIERWD-DANKVRQLM 284
Query: 468 SEAGMKKKPGCSIIEVGDEVEEFLAGD 494
G KK PGCS ++VGD++ FL G+
Sbjct: 283 KSRGAKKNPGCSWVKVGDKMHAFLVGE 203
>BP038410
Length = 468
Score = 70.9 bits (172), Expect = 5e-13
Identities = 41/130 (31%), Positives = 65/130 (49%)
Frame = -2
Query: 217 FKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATA 276
+ V LF +M G+ + S ++ +L A G G +H Y + S+ + TA
Sbjct: 392 WNSVFDLFTKMCWSGLIASASTVLILLPAAGDTGNFVVGKSLHGYYIQIGFSSHLNVLTA 213
Query: 277 LVDMYSKCGCVESALTVFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPT 336
L+DMY+K G V +F+ + DKD WN +I A +G V ++L L +M G +
Sbjct: 212 LIDMYAKTGLVHWGRKIFDSLVDKDVVLWNCLIGNYARNGLVGEALYLLQKMRIQGMKTN 33
Query: 337 ETTFVAVLTA 346
+TFV +L A
Sbjct: 32 SSTFVGLLLA 3
Score = 55.1 bits (131), Expect = 3e-08
Identities = 31/112 (27%), Positives = 55/112 (48%), Gaps = 1/112 (0%)
Frame = -2
Query: 86 CYSSMLRTGITVNNYTFPPLIKASLALLSSSSSSNFT-GRLVHGHVVKFGFHDDPYVVSG 144
C+S ++ + TV + L ++ + NF G+ +HG+ ++ GF V++
Sbjct: 359 CWSGLIASASTV-----------LILLPAAGDTGNFVVGKSLHGYYIQIGFSSHLNVLTA 213
Query: 145 FIEFYCASRQLQTARLLFDQTTEKDVVLWTAMIDGYGKVGDVESARELFEEM 196
I+ Y + + R +FD +KDVVLW +I Y + G V A L ++M
Sbjct: 212 LIDMYAKTGLVHWGRKIFDSLVDKDVVLWNCLIGNYARNGLVGEALYLLQKM 57
>BP063629
Length = 525
Score = 64.3 bits (155), Expect = 5e-11
Identities = 46/146 (31%), Positives = 71/146 (48%), Gaps = 6/146 (4%)
Frame = +1
Query: 8 SLVKKCKTLTQLKQVHAHILRCRRINHIAPLLSAAAATS------NFSYARSIFLHLTHR 61
SL+K CK++ +LKQ+ A I C + L+ A S +F YA IF H+
Sbjct: 106 SLLKSCKSMCELKQIQALIF-CSGLQQDRDTLNKLMAISTDSSIGDFHYALRIFDHIQQP 282
Query: 62 NTFIHNTMIRAYLLHAPSPAPALSCYSSMLRTGITVNNYTFPPLIKASLALLSSSSSSNF 121
+ F +N MI+A+ S A+S + + G+ +NYT+P ++KA L
Sbjct: 283 SLFNYNVMIKAFAKKG-SFRRAISLFQQLREDGVWPDNYTYPYVLKAIGCLGDVGQ---- 447
Query: 122 TGRLVHGHVVKFGFHDDPYVVSGFIE 147
GR VH V+K G D YV + ++
Sbjct: 448 -GRKVHAFVIKSGLEFDAYVCNSLMD 522
>TC20002 similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (4%)
Length = 560
Score = 45.4 bits (106), Expect = 2e-05
Identities = 37/140 (26%), Positives = 59/140 (41%), Gaps = 6/140 (4%)
Frame = -3
Query: 65 IHNTMIRAYLLHAPS-PAPALSCYSSMLRTGITVNNYTFPPLIKASLALLSSSSSSNFTG 123
I+ T Y+L S P P L+ S+ T + + L KA L++ + +
Sbjct: 489 INITFFLMYILILISKPKPPLNRRSATSTTAVCTMSPKDATLWKAQQTLMTLFNRCSTMN 310
Query: 124 RL--VHGHVVKFGFHDDPYVVSGFIEFYCAS---RQLQTARLLFDQTTEKDVVLWTAMID 178
L +H + + GFH + VV I F S + A +FD+ + D LW MI
Sbjct: 309 HLKEIHARIYQTGFHQNHLVVGKIIVFCAVSVPAGDMNYAVSVFDRVDKPDAFLWNTMIR 130
Query: 179 GYGKVGDVESARELFEEMPE 198
G+G E A ++ M +
Sbjct: 129 GFGNTNQPEKAVLFYKRMQQ 70
Score = 42.0 bits (97), Expect = 2e-04
Identities = 31/113 (27%), Positives = 53/113 (46%), Gaps = 8/113 (7%)
Frame = -3
Query: 3 QNNLTSLVKKCKTLTQLKQVHAHILRCR-RINH-----IAPLLSAAAATSNFSYARSIFL 56
Q L +L +C T+ LK++HA I + NH I + + + +YA S+F
Sbjct: 354 QQTLMTLFNRCSTMNHLKEIHARIYQTGFHQNHLVVGKIIVFCAVSVPAGDMNYAVSVFD 175
Query: 57 HLTHRNTFIHNTMIRAYLLHAPSPAPALSCYSSMLR--TGITVNNYTFPPLIK 107
+ + F+ NTMIR + + P A+ Y M + + + +TF L+K
Sbjct: 174 RVDKPDAFLWNTMIRGF-GNTNQPEKAVLFYKRMQQGEPHVVPDTFTFSFLLK 19
>TC15322 weakly similar to UP|Q9LG23 (Q9LG23) F14J16.14
(At1g55890/F14J16_4), partial (33%)
Length = 588
Score = 44.7 bits (104), Expect = 4e-05
Identities = 27/75 (36%), Positives = 45/75 (60%), Gaps = 5/75 (6%)
Frame = +2
Query: 176 MIDGYGKVGDVESARELFEEMPERN----AVSWSAMMAAYSRVSDFKQVLALFEEMQNR- 230
+I YGK G + AR++F+EMPERN +S++A++AAY ++ V +F+E+
Sbjct: 359 LITLYGKSGMPKRARQMFDEMPERNCTRTVLSFNALLAAYLHSKQYRVVERIFKEVPGEI 538
Query: 231 GMKPNDSVLVTVLTA 245
+KP+ TV+ A
Sbjct: 539 SVKPDLVSYNTVIKA 583
>TC19070 weakly similar to UP|Q9LRP6 (Q9LRP6) DNA-binding protein
(AT3g15590/MQD17_5), partial (20%)
Length = 744
Score = 43.1 bits (100), Expect = 1e-04
Identities = 43/162 (26%), Positives = 73/162 (44%), Gaps = 9/162 (5%)
Frame = +3
Query: 206 AMMAAYSRVSDFKQVLALFEEMQNRGM--KPNDSVLVTVLTACAHLGALTQG--LWVHSY 261
A + A+ R+ ++ A+FE M N+ N SVL+ V A+ L +G L
Sbjct: 18 AAVEAWGRLQKIEEAEAIFEMMANKWKLSSKNYSVLLKVY---ANHKMLKKGKDLIQRMG 188
Query: 262 AKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADKDA-----GAWNAMISGVALSG 316
CK+ P+ +LV +Y++ G VE A V + + ++ + A++ A G
Sbjct: 189 ENGCKI--GPLTWDSLVKLYTQAGEVEKADAVLQKVMEQSRMTPMFNTYMAVMEQYAKRG 362
Query: 317 DVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLR 358
DV S ++FH+M G T F ++ A + G+R
Sbjct: 363 DVHNSEKIFHRMRQAGYTSRITPFQVLVQAYIKAKVPAYGIR 488
>AI967878
Length = 393
Score = 42.7 bits (99), Expect = 1e-04
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 20/88 (22%)
Frame = +1
Query: 304 AWNAMISGVALSGDVRKSLELFHQM-----------------AAHGT---QPTETTFVAV 343
+WNAMI G + G+ L LF +M A G P E TF+ +
Sbjct: 43 SWNAMILGHCIHGNPEDGLSLFDEMVGMDKVKGEVEIDESPCADRGVVRLPPDEVTFIGI 222
Query: 344 LTACTHGGMVHQGLRLFEEMSCTYGVEP 371
L AC ++ +G F++M+ +G++P
Sbjct: 223 LCACARAELLAEGRSYFKQMTDVFGLKP 306
Score = 29.6 bits (65), Expect = 1.2
Identities = 27/99 (27%), Positives = 43/99 (43%)
Frame = +1
Query: 166 TEKDVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFE 225
T +++V W AMI G+ G+ E LF+EM + V + S +D V
Sbjct: 25 TNRNLVSWNAMILGHCIHGNPEDGLSLFDEMVGMDKVKGEVEIDE-SPCADRGVV----- 186
Query: 226 EMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKR 264
+ P++ + +L ACA L +G SY K+
Sbjct: 187 -----RLPPDEVTFIGILCACARAELLAEG---RSYFKQ 279
>AV409405
Length = 424
Score = 42.0 bits (97), Expect = 2e-04
Identities = 25/85 (29%), Positives = 37/85 (43%)
Frame = +1
Query: 112 LLSSSSSSNFTGRLVHGHVVKFGFHDDPYVVSGFIEFYCASRQLQTARLLFDQTTEKDVV 171
LL + T +L H +K G D ++V+ Y + A LFD+T +K V
Sbjct: 163 LLETCHCEKSTSQL-HSLCLKLGLTHDSFIVTKLNVLYGKHASIHHAHKLFDETPDKSVY 339
Query: 172 LWTAMIDGYGKVGDVESARELFEEM 196
LW A++ Y G+ LF M
Sbjct: 340 LWNALLRRYRAEGEWAETLSLFRRM 414
Score = 40.0 bits (92), Expect = 0.001
Identities = 15/48 (31%), Positives = 28/48 (58%)
Frame = +1
Query: 180 YGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEM 227
YGK + A +LF+E P+++ W+A++ Y ++ + L+LF M
Sbjct: 271 YGKHASIHHAHKLFDETPDKSVYLWNALLRRYRAEGEWAETLSLFRRM 414
>AV767495
Length = 586
Score = 40.8 bits (94), Expect = 5e-04
Identities = 48/201 (23%), Positives = 85/201 (41%), Gaps = 4/201 (1%)
Frame = +2
Query: 167 EKDVVLWTAMIDGYGKVGDVESARELFEEMPER----NAVSWSAMMAAYSRVSDFKQVLA 222
E DVV ++A+ID K G VES+ L + M E+ N V++++++ A+ ++S + +
Sbjct: 20 EADVVFYSALIDALCKNGLVESSMVLLDAMIEKGIRPNVVTYNSIIDAFGQLSALECGVD 199
Query: 223 LFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYS 282
+ + P+ S+L+ GAL LA D
Sbjct: 200 TSVQANEHRVVPSSSMLID--------GALQN------------------LAIGKED--- 292
Query: 283 KCGCVESALTVFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVA 342
+ + +FE +A + +G + G S D L LF +M +P TF A
Sbjct: 293 -----DRIMKMFEQLAAEKSGQIKKDMRG---SQDKFCILWLFRKMHEMEIKPNVVTFSA 448
Query: 343 VLTACTHGGMVHQGLRLFEEM 363
+L AC++ +L +E+
Sbjct: 449 ILNACSNCKSFEDASKLLDEL 511
Score = 28.1 bits (61), Expect = 3.6
Identities = 33/178 (18%), Positives = 79/178 (43%), Gaps = 11/178 (6%)
Frame = +2
Query: 263 KRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADK----DAGAWNAMISGVALSGDV 318
K+ +++++ + +AL+D K G VES++ + + + +K + +N++I +
Sbjct: 5 KQERLEADVVFYSALIDALCKNGLVESSMVLLDAMIEKGIRPNVVTYNSIIDAFGQLSAL 184
Query: 319 RKSLELFHQMAAHGTQPTETTFV-AVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYA 377
++ Q H P+ + + L G + +++FE+++ E G+
Sbjct: 185 ECGVDTSVQANEHRVVPSSSMLIDGALQNLAIGKEDDRIMKMFEQLA----AEKSGQ--- 343
Query: 378 CVVDLLSRAGMVKEAEKF-----MEKMGGL-VEADLNVWGAILNACRIYKNVDVGNRV 429
+ M +KF KM + ++ ++ + AILNAC K+ + +++
Sbjct: 344 ------IKKDMRGSQDKFCILWLFRKMHEMEIKPNVVTFSAILNACSNCKSFEDASKL 499
>AV766731
Length = 576
Score = 38.5 bits (88), Expect = 0.003
Identities = 28/116 (24%), Positives = 52/116 (44%), Gaps = 8/116 (6%)
Frame = -3
Query: 138 DPYVVSGFIEFYCASRQLQTARLLF----DQTTEKDVVLWTAMIDGYGKVGDVESARELF 193
D + + C L A LF D + ++ + +IDG KVG ++ A+E+F
Sbjct: 445 DLITTNSLFDGLCKHHLLDKAIALFMKVKDPKIQPNIPPYPVIIDGLCKVGRLKIAQEIF 266
Query: 194 E----EMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTA 245
+ E N ++++ M+ Y + + AL +M++ G PN T++ A
Sbjct: 265 QVLLSEGYNLNVMTYTVMINGYCKEGLLDEAQALLSKMEDNGCIPNAVNFQTIICA 98
>AV769479
Length = 287
Score = 37.7 bits (86), Expect = 0.005
Identities = 19/74 (25%), Positives = 37/74 (49%)
Frame = +1
Query: 123 GRLVHGHVVKFGFHDDPYVVSGFIEFYCASRQLQTARLLFDQTTEKDVVLWTAMIDGYGK 182
G+ +H H++K + + + F+ FY ++ +A FD+ ++DVV WT +I +
Sbjct: 52 GKQIHAHILKSKWRN-LIADNAFVNFYAECGKISSAFRTFDRMAKRDVVCWTTIITACSQ 228
Query: 183 VGDVESARELFEEM 196
G A + +M
Sbjct: 229 QGLGHEALLILSQM 270
>AV779633
Length = 440
Score = 36.6 bits (83), Expect = 0.010
Identities = 30/139 (21%), Positives = 64/139 (45%), Gaps = 29/139 (20%)
Frame = -2
Query: 134 GFHDDPYVVSGFIEFYCASRQLQTARLLFDQT----TEKDVVLWTAMIDGYGKVGDVESA 189
G D ++ I+ YC + + A L + + + D+V + +++ G+ K GD+ A
Sbjct: 430 GIARDVIGLNTLIDGYCEAGLMSQALALMENSWKTGVKPDIVSYNSLLKGFCKAGDLVRA 251
Query: 190 RELFEEM-----------PERNAV--------------SWSAMMAAYSRVSDFKQVLALF 224
LF+E+ + NAV +++ +++AY + ++ +L+
Sbjct: 250 ESLFDEILGFQRDGESGQLKNNAVDTRDELRNIRPTLATYTTLISAYGKHCGIEESRSLY 71
Query: 225 EEMQNRGMKPNDSVLVTVL 243
E+M G+ P+ VL+ +L
Sbjct: 70 EQMVMSGIMPD--VLLVIL 20
Score = 32.3 bits (72), Expect = 0.19
Identities = 18/70 (25%), Positives = 36/70 (50%), Gaps = 4/70 (5%)
Frame = -2
Query: 162 FDQTTEKDVVLWTAMIDGYGKVGDVESARELFEEM----PERNAVSWSAMMAAYSRVSDF 217
FD +DV+ +IDGY + G + A L E + + VS+++++ + + D
Sbjct: 439 FDGGIARDVIGLNTLIDGYCEAGLMSQALALMENSWKTGVKPDIVSYNSLLKGFCKAGDL 260
Query: 218 KQVLALFEEM 227
+ +LF+E+
Sbjct: 259 VRAESLFDEI 230
>TC19836
Length = 464
Score = 35.0 bits (79), Expect = 0.030
Identities = 17/78 (21%), Positives = 39/78 (49%)
Frame = +2
Query: 409 VWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYREAGWDAEAKRVRSMVS 468
+W ++++AC+++ +D+ + ++I + + L +Y E G ++V +
Sbjct: 32 IWSSLVSACKLHGRLDIAEMLGPQLIRSEPDNAANYTLLNMIYAEHGHWLHKEQVMEAMK 211
Query: 469 EAGMKKKPGCSIIEVGDE 486
+KK G S IE G++
Sbjct: 212 LQRLKKCYGFSRIETGNQ 265
>BP039174
Length = 564
Score = 31.2 bits (69), Expect = 0.43
Identities = 19/88 (21%), Positives = 40/88 (44%)
Frame = -2
Query: 204 WSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAK 263
+ A++ AYSR+ + + +F+ +Q G+ P+D + V+ A G + K
Sbjct: 551 YKALLRAYSRIGNAEGAQRVFDAIQLAGIIPDDKICGLVIKAYGMAGQSEKARIAFENMK 372
Query: 264 RCKVDSNPILATALVDMYSKCGCVESAL 291
R ++ +++ Y K + +AL
Sbjct: 371 RAGIEPTDRCIGSVLVAYEKESKLNTAL 288
>TC14645 similar to UP|Q41122 (Q41122) Proline-rich protein precursor,
partial (52%)
Length = 1060
Score = 30.4 bits (67), Expect = 0.73
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Frame = +2
Query: 76 HAPSPAPALSCYSSMLRTGITVNNYTFPPLIKASL--ALLSSSSSSNFTG 123
H PSPAPA + Y+++ + + + F P ++A L +LS S +N G
Sbjct: 398 HPPSPAPAPALYTTITLPPLLLFTHLFTPSLEALLLFKVLSLPSPANMQG 547
>BF177625
Length = 284
Score = 30.4 bits (67), Expect = 0.73
Identities = 19/80 (23%), Positives = 40/80 (49%), Gaps = 9/80 (11%)
Frame = +1
Query: 150 CASRQLQTARL--LFDQTTEK----DVVLWTAMIDGYGKVGDVESARELFEEMPER---N 200
C S+ L L LFD+ ++ + + + M+D +GK R L+ ++ +
Sbjct: 34 CCSQALPVDELSRLFDEMLQRGFAPNTITYNVMLDVFGKAKLFRKVRRLYFMAKKQGLVD 213
Query: 201 AVSWSAMMAAYSRVSDFKQV 220
++++ ++AAY + DFK +
Sbjct: 214 VITYNTIIAAYGKNKDFKNM 273
>AV772727
Length = 528
Score = 29.6 bits (65), Expect = 1.2
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 4/55 (7%)
Frame = -2
Query: 146 IEFYCASRQLQTARLLFDQTTEK----DVVLWTAMIDGYGKVGDVESARELFEEM 196
++ C + + A L ++ +K D+ + +IDG K G V+ A+ELF+++
Sbjct: 506 LDALCKNHHVDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVKDAQELFQDL 342
>BP035488
Length = 534
Score = 29.3 bits (64), Expect = 1.6
Identities = 14/49 (28%), Positives = 22/49 (44%)
Frame = +3
Query: 406 DLNVWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYREA 454
D+ W ILN + N RVWK ++ + C + TY + +A
Sbjct: 66 DIKTWNVILNGWCVLGNAHEAKRVWKDIM---ASKCRPDLFTYATFIKA 203
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,358,536
Number of Sequences: 28460
Number of extensions: 137453
Number of successful extensions: 689
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 663
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 680
length of query: 519
length of database: 4,897,600
effective HSP length: 94
effective length of query: 425
effective length of database: 2,222,360
effective search space: 944503000
effective search space used: 944503000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0252d.2


