

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.3
(282 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
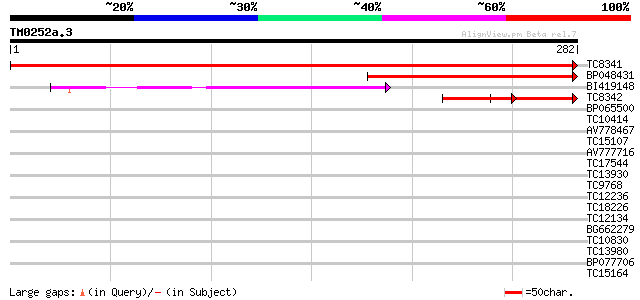
Score E
Sequences producing significant alignments: (bits) Value
TC8341 similar to UP|Q94FX1 (Q94FX1) Heme oxygenase 1 (Fragment)... 573 e-164
BP048431 170 2e-43
BI419148 130 2e-31
TC8342 homologue to UP|Q94FX0 (Q94FX0) Heme oxygenase 3 (Fragmen... 77 4e-15
BP065500 39 0.001
TC10414 similar to GB|AAP13363.1|30023660|BT006255 At5g55710 {Ar... 32 0.093
AV778467 30 0.35
TC15107 similar to UP|Q8LJP8 (Q8LJP8) Dehydroascorbate reductase... 30 0.35
AV777716 28 1.3
TC17544 similar to PIR|T04919|T04919 DNA-binding protein homolog... 27 3.0
TC13930 similar to UP|Q7PPK1 (Q7PPK1) ENSANGP00000005139 (Fragme... 27 3.9
TC9768 similar to GB|AAP68290.1|31711868|BT008851 At3g12156 {Ara... 27 3.9
TC12236 similar to UP|Q9LJE2 (Q9LJE2) Lysyl-tRNA synthetase, par... 27 3.9
TC18226 similar to UP|Q944G3 (Q944G3) Acetyl Co-A acetyltransfer... 27 5.1
TC12134 UP|IM23_DROME (Q9V8F5) Immune-induced peptide 23 precurs... 23 5.7
BG662279 26 6.7
TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, ... 26 6.7
TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.3... 26 6.7
BP077706 26 8.7
TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, p... 26 8.7
>TC8341 similar to UP|Q94FX1 (Q94FX1) Heme oxygenase 1 (Fragment), partial
(86%)
Length = 1286
Score = 573 bits (1477), Expect = e-164
Identities = 282/282 (100%), Positives = 282/282 (100%)
Frame = +2
Query: 1 MASATVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAA 60
MASATVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAA
Sbjct: 242 MASATVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAA 421
Query: 61 ETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFL 120
ETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFL
Sbjct: 422 ETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFL 601
Query: 121 VDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNY 180
VDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNY
Sbjct: 602 VDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNY 781
Query: 181 AQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLL 240
AQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLL
Sbjct: 782 AQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLL 961
Query: 241 QNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
QNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS
Sbjct: 962 QNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 1087
>BP048431
Length = 444
Score = 170 bits (431), Expect = 2e-43
Identities = 82/104 (78%), Positives = 92/104 (87%)
Frame = -2
Query: 179 NYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQ 238
NYA YLR+LS +DP A ICHFY+IYFA SAGGRMIG+KIAGELL+++GL FYKWDGD S
Sbjct: 443 NYAPYLRELSPDDPPALICHFYHIYFAPSAGGRMIGEKIAGELLHHQGLGFYKWDGDPSP 264
Query: 239 LLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
LL NVRD+L++VAE WTREEKN CL ETEKSFKWSGEILRLILS
Sbjct: 263 LLPNVRDQLDQVAEPWTREEKNPCLGETEKSFKWSGEILRLILS 132
>BI419148
Length = 537
Score = 130 bits (328), Expect = 2e-31
Identities = 71/178 (39%), Positives = 101/178 (55%), Gaps = 9/178 (5%)
Frame = +2
Query: 21 SPPPPPHR---------QFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGES 71
+PPPPP + + R ++ T+ +Q+ R R RK YPGE+
Sbjct: 56 TPPPPPSKWSSATRTTVKMRVVFNNTSPPVQKKRKRY---------------RKLYPGET 190
Query: 72 KGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLE 131
G EEMRFVAMKL T P W+P++DG+L+FLVD+ +V+ TLE
Sbjct: 191 TGITEEMRFVAMKLRNDTTV------ATAPSTSHDDTWQPSMDGFLRFLVDNNLVFATLE 352
Query: 132 KIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQ 189
+IV ++ + SYA R GLERS + KDL WF+E+G +P PSSPG+ YA+YL +L++
Sbjct: 353 RIVDESDNVSYAYMRKCGLERSKGILKDLAWFEERGLVVPTPSSPGVTYAKYLEELAE 526
>TC8342 homologue to UP|Q94FX0 (Q94FX0) Heme oxygenase 3 (Fragment),
partial (25%)
Length = 570
Score = 76.6 bits (187), Expect = 4e-15
Identities = 36/37 (97%), Positives = 37/37 (99%)
Frame = +2
Query: 216 KIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAE 252
+IAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAE
Sbjct: 53 QIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAE 163
Score = 67.4 bits (163), Expect = 3e-12
Identities = 31/43 (72%), Positives = 35/43 (81%)
Frame = +1
Query: 240 LQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
L N+ ++ + WTREEKNHCLEETEKSFKWSGEILRLILS
Sbjct: 316 LMNIPFGISLFYQPWTREEKNHCLEETEKSFKWSGEILRLILS 444
>BP065500
Length = 512
Score = 38.5 bits (88), Expect = 0.001
Identities = 23/37 (62%), Positives = 27/37 (72%)
Frame = -2
Query: 244 RDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLI 280
RDK K ++WT EEKNHCL E EKSF++ EIL LI
Sbjct: 265 RDK*KK--KEWTWEEKNHCL-EVEKSFQF*REIL*LI 164
>TC10414 similar to GB|AAP13363.1|30023660|BT006255 At5g55710 {Arabidopsis
thaliana;}, partial (34%)
Length = 504
Score = 32.3 bits (72), Expect = 0.093
Identities = 14/44 (31%), Positives = 25/44 (56%)
Frame = +1
Query: 22 PPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARK 65
PPPPPH++ RS RL ++ R +++++++ T RK
Sbjct: 238 PPPPPHQEARSSQIQRQRLRRRPRPFDLRALLLLPILRRHTVRK 369
>AV778467
Length = 491
Score = 30.4 bits (67), Expect = 0.35
Identities = 19/48 (39%), Positives = 24/48 (49%), Gaps = 4/48 (8%)
Frame = +1
Query: 221 LLNNKGLEFYKWDGDLSQLLQNVRDKLNKVA----EQWTREEKNHCLE 264
LL NK E SQ LQN R KLN+V QWT +++ C +
Sbjct: 166 LLTNKAYE--------SQKLQNKRKKLNRVGHGLKHQWTIRDRDRCFK 285
>TC15107 similar to UP|Q8LJP8 (Q8LJP8) Dehydroascorbate reductase, partial
(98%)
Length = 1132
Score = 30.4 bits (67), Expect = 0.35
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Frame = +3
Query: 6 VVSQIQSLYIIKPRLSPPPPP-HRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETAR 64
V SQ + ++ R PPPPP HR+F T+LL QH F++ V V +++ T R
Sbjct: 60 VHSQSSRVGALRHRQPPPPPPNHRRF-------TQLL-QHPFQEASQGVHVFSSSFGTRR 215
Query: 65 KR 66
R
Sbjct: 216 NR 221
>AV777716
Length = 494
Score = 28.5 bits (62), Expect = 1.3
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 4/48 (8%)
Frame = +2
Query: 221 LLNNKGLEFYKWDGDLSQLLQNVRDKLNKVA----EQWTREEKNHCLE 264
LL NK E SQ LQN R KLN V QWT ++ + C +
Sbjct: 95 LLTNKAYE--------SQKLQNKRKKLNNVCHCLKHQWTIKDLDRCFK 214
>TC17544 similar to PIR|T04919|T04919 DNA-binding protein homolog T9A21.10 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(14%)
Length = 578
Score = 27.3 bits (59), Expect = 3.0
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -1
Query: 2 ASATVVSQIQSLYIIKPRLSPPPPP 26
AS ++I L ++ R+SPPPPP
Sbjct: 398 ASEDEEAEIDELRVVVARVSPPPPP 324
>TC13930 similar to UP|Q7PPK1 (Q7PPK1) ENSANGP00000005139 (Fragment),
partial (8%)
Length = 570
Score = 26.9 bits (58), Expect = 3.9
Identities = 20/62 (32%), Positives = 31/62 (49%), Gaps = 1/62 (1%)
Frame = +3
Query: 6 VVSQIQSLYIIKPR-LSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETAR 64
++S SL +I+ + L PPPPP +F + P + RF + S + S+ A TA
Sbjct: 219 LLSNTTSLGLIRSK*LPPPPPPPPRFATPITPRPQFPSPCRF-TLDSQGLESSVTAVTAA 395
Query: 65 KR 66
R
Sbjct: 396 SR 401
>TC9768 similar to GB|AAP68290.1|31711868|BT008851 At3g12156 {Arabidopsis
thaliana;}, partial (51%)
Length = 865
Score = 26.9 bits (58), Expect = 3.9
Identities = 11/25 (44%), Positives = 13/25 (52%)
Frame = +2
Query: 249 KVAEQWTREEKNHCLEETEKSFKWS 273
K+A WT E HC T KS W+
Sbjct: 704 KIASWWTISEGKHCNHGT*KSILWT 778
>TC12236 similar to UP|Q9LJE2 (Q9LJE2) Lysyl-tRNA synthetase, partial (16%)
Length = 429
Score = 26.9 bits (58), Expect = 3.9
Identities = 15/54 (27%), Positives = 23/54 (41%), Gaps = 9/54 (16%)
Frame = +3
Query: 7 VSQIQSLYIIKPRLSPPPPPH---------RQFRSIYFPTTRLLQQHRFRQMKS 51
V + + P L PPPPPH R+ R + P R + R++K+
Sbjct: 75 VEHLAGAFSAHPLLQPPPPPHPPVKTGGALRRPRRLQLPIERPFEPFA*RRLKN 236
>TC18226 similar to UP|Q944G3 (Q944G3) Acetyl Co-A acetyltransferase,
partial (19%)
Length = 426
Score = 26.6 bits (57), Expect = 5.1
Identities = 16/41 (39%), Positives = 20/41 (48%)
Frame = -1
Query: 17 KPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSA 57
K R PPPP ++ + YFP RFR KSV + A
Sbjct: 228 KTRAEAPPPP*QEAPTPYFPWL------RFRTPKSVTRIRA 124
>TC12134 UP|IM23_DROME (Q9V8F5) Immune-induced peptide 23 precursor
(DIM-23), partial (9%)
Length = 434
Score = 22.7 bits (47), Expect(2) = 5.7
Identities = 9/13 (69%), Positives = 10/13 (76%)
Frame = +3
Query: 21 SPPPPPHRQFRSI 33
+PPPPP FRSI
Sbjct: 393 APPPPPPP*FRSI 431
Score = 21.9 bits (45), Expect(2) = 5.7
Identities = 12/25 (48%), Positives = 15/25 (60%), Gaps = 2/25 (8%)
Frame = +2
Query: 4 ATVVSQIQSLYIIKPRL--SPPPPP 26
+T +S L I+ P L SPPPPP
Sbjct: 335 STFLSICLKLSILDPFLVLSPPPPP 409
>BG662279
Length = 337
Score = 26.2 bits (56), Expect = 6.7
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +2
Query: 16 IKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQ 48
I+PR PP P HR +++ P + LQ H R+
Sbjct: 119 IQPRFGPPHPHHRARQAL--PPSPQLQSHHPRR 211
>TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, partial
(24%)
Length = 702
Score = 26.2 bits (56), Expect = 6.7
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Frame = +1
Query: 2 ASATVVSQIQSLYIIKPRLSPPPPPHRQFRSIY---FPTTRLLQQHRFRQMK 50
A+ T S + +L+ + P PPPH + FP T LLQ H FR ++
Sbjct: 106 ATTTTTSSLITLHFLTHL--PLPPPHSTSLHSHQPRFPPTNLLQSH-FRHLQ 252
>TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.33), partial
(12%)
Length = 565
Score = 26.2 bits (56), Expect = 6.7
Identities = 11/27 (40%), Positives = 12/27 (43%)
Frame = +3
Query: 18 PRLSPPPPPHRQFRSIYFPTTRLLQQH 44
P LS P H S+YFP L H
Sbjct: 105 PTLSSYTPKHTSINSLYFPLPHFLSLH 185
>BP077706
Length = 422
Score = 25.8 bits (55), Expect = 8.7
Identities = 12/28 (42%), Positives = 16/28 (56%), Gaps = 5/28 (17%)
Frame = +1
Query: 13 LYIIKPRLSP-----PPPPHRQFRSIYF 35
++IIK SP PPPPH + I+F
Sbjct: 55 IFIIKGPRSPQPATSPPPPHPNLKPIFF 138
>TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, partial
(57%)
Length = 1185
Score = 25.8 bits (55), Expect = 8.7
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = -3
Query: 18 PRLSPPPPPHRQFRS 32
PR++PPPPP F S
Sbjct: 838 PRIAPPPPPAPAFHS 794
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,923,778
Number of Sequences: 28460
Number of extensions: 67774
Number of successful extensions: 945
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 879
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 928
length of query: 282
length of database: 4,897,600
effective HSP length: 89
effective length of query: 193
effective length of database: 2,364,660
effective search space: 456379380
effective search space used: 456379380
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0252a.3


