

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.1
(346 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
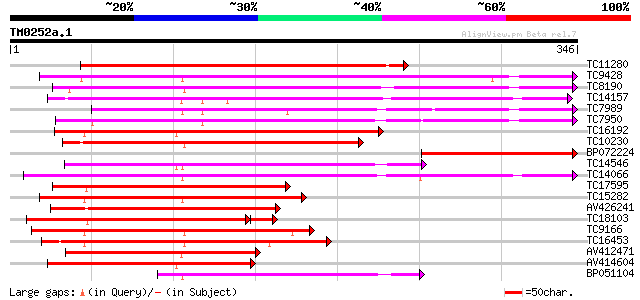
Score E
Sequences producing significant alignments: (bits) Value
TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), p... 271 9e-74
TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), pa... 243 4e-65
TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 p... 224 1e-59
TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , p... 217 3e-57
TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , par... 216 4e-57
TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), part... 209 7e-55
TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%) 192 5e-50
TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precurso... 189 4e-49
BP072224 187 2e-48
TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxida... 179 5e-46
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 168 1e-42
TC17595 similar to UP|Q9XFI7 (Q9XFI7) Peroxidase (Fragment), par... 159 9e-40
TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fra... 157 3e-39
AV426241 153 4e-38
TC18103 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 ... 141 3e-36
TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%) 140 4e-34
TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, part... 128 1e-30
AV412471 127 2e-30
AV414604 125 8e-30
BP051104 124 3e-29
>TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), partial
(68%)
Length = 614
Score = 271 bits (694), Expect = 9e-74
Identities = 130/200 (65%), Positives = 158/200 (79%)
Frame = +3
Query: 44 QAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGS 103
Q + GFY +CP AE+ VRST+ES+ N DPT+A L+R+HFHDCFVQGCD SVLI G+
Sbjct: 15 QGGSRVGFYLGTCPRAESIVRSTVESHVNSDPTLAAGLLRMHFHDCFVQGCDASVLIAGA 194
Query: 104 SAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTG 163
ER A+ NLSLRGFEVI+DAK+++EA CPGVVSCADILA+AARD+V LS G SW VPTG
Sbjct: 195 GTERTAIPNLSLRGFEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTG 374
Query: 164 RRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRL 223
RRDGR+S ++ +NLP+P D V VQ+QKF AKGL+ DLVTLVG HTIG T C+FFS RL
Sbjct: 375 RRDGRVSQASDVNNLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGGHTIGTTACQFFSNRL 554
Query: 224 YNFTTTGNSDPTINQATLAQ 243
YNFT+ G DP+I+ + L Q
Sbjct: 555 YNFTSNG-PDPSIDASFLLQ 611
>TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1354
Score = 243 bits (619), Expect = 4e-65
Identities = 137/338 (40%), Positives = 194/338 (56%), Gaps = 10/338 (2%)
Frame = +2
Query: 19 YKVSSQMEAMLLGSLMILMISFII---AQAQLKTGFYSSSCPTAEATVRSTIESYFNKDP 75
Y +M + L + ++I ++ + AQL FY +CP + VR + + DP
Sbjct: 23 YLSEEKMSPIHLAVVFCVVIGALVPYSSDAQLDNSFYKDTCPNVHSIVREVVRNVSKSDP 202
Query: 76 TIAPSLIRLHFHDCFVQGCDGSVLIVGSS---AERNALANL-SLRGFEVIEDAKSQLEAI 131
+ SLIRLHFHDCFVQGCD S+L+ +S +E+ A N S+RG +V+ K+ +E
Sbjct: 203 RMLGSLIRLHFHDCFVQGCDASILLNDTSTIVSEQGAFPNNNSIRGLDVVNQIKTAVENA 382
Query: 132 CPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQAS-NLPSPLDPVSVQRQ 190
CP VSCADILA+AA + L+NG W VP GRRD ++ + A+ NLP P +S
Sbjct: 383 CPNTVSCADILALAAEISSVLANGVDWKVPLGRRDSLTANRSLANQNLPGPNFNLSQLNA 562
Query: 191 KFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPK 250
FA + L+ DLV L GAHTIG+ +C+FF RLYNF+ TGN DPT+N L +++CP
Sbjct: 563 SFAVQNLNITDLVALSGAHTIGRGQCQFFVNRLYNFSNTGNPDPTLNTTYLQTLRSICPH 742
Query: 251 NGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDS--ATQSVVQSYAGNIRG 308
G G LD +P D +F N++ G G+ +SD L+ S AT ++V S++ N
Sbjct: 743 GGPGTTLAHLDPTTPDTLDSQYFSNLQGGKGLFQSDPVLFSTSGAATVAIVNSFSSN--- 913
Query: 309 LLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
F F +MIK+S I V T ++GEIRK C+ N
Sbjct: 914 --PTLFFENFKASMIKMSRIGVLTGSQGEIRKPCNFVN 1021
>TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (96%)
Length = 1202
Score = 224 bits (572), Expect = 1e-59
Identities = 131/329 (39%), Positives = 184/329 (55%), Gaps = 9/329 (2%)
Frame = +1
Query: 27 AMLLGSLMI----LMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLI 82
AM+L S+ + L I AQL + FY+ +CP AT+++ + K+ + SL+
Sbjct: 70 AMILPSMEVRFFFLFCLIGIGSAQLSSTFYAKTCPLVLATIKTQVNLAVAKEARMGASLL 249
Query: 83 RLHFHDCFVQGCDGSVLIVGSSA---ERNALANL-SLRGFEVIEDAKSQLEAICPGVVSC 138
RLHFHDCFVQGCD S+L+ +S+ E+ A N S+RG++VI+ KS++E++CPGVVSC
Sbjct: 250 RLHFHDCFVQGCDASILLDDTSSFTGEKTAGPNANSVRGYDVIDTIKSKVESLCPGVVSC 429
Query: 139 ADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGL 197
ADI+AVAARD+V G SW+VP GRRD +S + A S LP P + F+ KG
Sbjct: 430 ADIVAVAARDSVVALGGFSWAVPLGRRDSTTASLSSANSELPGPSSNLDGLNTAFSNKGF 609
Query: 198 DDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRK 257
++V L G+HTIGQ C FF R+Y T I+ Q CP NG
Sbjct: 610 TTREMVALSGSHTIGQARCLFFRTRIYYET-------HIDSTFAKNLQGNCPFNGGDSNL 768
Query: 258 VSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYE 317
LD+ SP FD +++N++ G+ SDQ L+ +T S V SY N F +
Sbjct: 769 SPLDTTSPTTFDNGYYRNLQSQKGLFHSDQVLFNGGSTDSQVNSYVTN-----PASFKTD 933
Query: 318 FPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
F AM+K+ N+ T + G+IR C K N
Sbjct: 934 FANAMVKMGNLSPLTGSSGQIRTHCRKTN 1020
>TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , partial
(96%)
Length = 1390
Score = 217 bits (552), Expect = 3e-57
Identities = 127/328 (38%), Positives = 188/328 (56%), Gaps = 8/328 (2%)
Frame = +1
Query: 24 QMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIR 83
Q+ +L+ SL L + F AQL Y+S+CP ++ V+ ++ F + P+ +R
Sbjct: 61 QINRVLVLSLS-LTLCFYTCFAQLSPNHYASTCPNLQSIVKGVVQKKFQQTFVTVPATLR 237
Query: 84 LHFHDCFVQGCDGSVLIVGS---SAERNALANLSLR--GFEVIEDAKSQLEAI--CPGVV 136
L FHDCFVQGCD SV++ S AE++ NLSL GF+ + AK+ ++A+ C V
Sbjct: 238 LFFHDCFVQGCDASVMVASSGNNKAEKDHPDNLSLAGDGFDTVIKAKAAVDAVPQCRNKV 417
Query: 137 SCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQAS-NLPSPLDPVSVQRQKFAAK 195
SCADILA+A RD V L+ GPS++V GR DG +S ++ + LP P ++ FA++
Sbjct: 418 SCADILALATRDVVVLAGGPSYTVELGRFDGLVSRASDVNGRLPEPNFNLNQLNSLFASQ 597
Query: 196 GLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGL 255
GL D++ L GAHT+G + C FS R+Y+ DPT+N+ Q Q +CPKN N
Sbjct: 598 GLTQTDMIALSGAHTLGFSHCNRFSNRIYSTPV----DPTLNRNYATQLQQMCPKNVNPQ 765
Query: 256 RKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFA 315
+++D +P FD ++KN++ G G+ SDQ L+ D +++ V S+A N F
Sbjct: 766 IAINMDPTTPRTFDNIYYKNLQQGKGLFTSDQILFTDQRSKATVNSFASNSN-----TFN 930
Query: 316 YEFPKAMIKLSNIEVKTDTEGEIRKVCS 343
F AMIKL + VKT G+IR CS
Sbjct: 931 ANFAAAMIKLGRVGVKTARNGKIRTDCS 1014
>TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (85%)
Length = 1352
Score = 216 bits (550), Expect = 4e-57
Identities = 127/303 (41%), Positives = 176/303 (57%), Gaps = 7/303 (2%)
Frame = +3
Query: 51 FYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSS---AER 107
FY SSCP E+ +R ++ F+KD A L+RLHFHDCFVQGCD SVL+ GS+ +E+
Sbjct: 150 FYDSSCPKLESIIRKELKKVFDKDIAQAAGLLRLHFHDCFVQGCDASVLLDGSASGPSEK 329
Query: 108 NALANLSLR--GFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRR 165
+A NL+LR F++IED +SQ+E C VVSCADI A+AARDAV+LS GP + +P GRR
Sbjct: 330 DAPPNLTLRAQAFKIIEDLRSQIEKKCGRVVSCADITAIAARDAVFLSGGPDYELPLGRR 509
Query: 166 DGR--ISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRL 223
DG + + NLP+P + A K LD D+V+L G HTIG + C F+ RL
Sbjct: 510 DGLNFATRNVTLDNLPAPQSNTTTILNSLATKNLDPTDVVSLSGGHTIGISHCSSFTDRL 689
Query: 224 YNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVL 283
Y + DP ++Q + CP + V LD SP FD ++ ++ + G+
Sbjct: 690 Y-----PSKDPVMDQTFEKNLKLTCPASNTDNTTV-LDLRSPNTFDNKYYVDLMNRQGLF 851
Query: 284 ESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCS 343
SDQ L+ D T+ +V S+A N F +F AM+K+ + V T ++GEIR CS
Sbjct: 852 FSDQDLYTDKRTKDIVTSFAVN-----QSLFFEKFVVAMLKMGQLNVLTGSQGEIRANCS 1016
Query: 344 KFN 346
N
Sbjct: 1017VRN 1025
>TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), partial (86%)
Length = 1286
Score = 209 bits (531), Expect = 7e-55
Identities = 123/328 (37%), Positives = 178/328 (53%), Gaps = 10/328 (3%)
Frame = +1
Query: 29 LLGSLMILMISFIIAQAQLKT--------GFYSSSCPTAEATVRSTIESYFNKDPTIAPS 80
L+ S++ F+ ++AQ K FYS +CP E VR+ ++ KD AP
Sbjct: 28 LIFSILFTSHFFLGSEAQTKPPVVEGLSFSFYSKTCPKLETVVRNHLKKVLKKDNGQAPG 207
Query: 81 LIRLHFHDCFVQGCDGSVLIVGSSAERNALANLSLR--GFEVIEDAKSQLEAICPGVVSC 138
L+R+ FHDCFVQGCDGSVL+ GS ER+ AN+ +R + IED ++ + C +VSC
Sbjct: 208 LLRIFFHDCFVQGCDGSVLLDGSPGERDQPANIGIRPEALQTIEDIRALVHKQCGKIVSC 387
Query: 139 ADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLD 198
ADI +A+RDAV+L+ GP ++VP GRRDG S+ LPSP++ + + FA + D
Sbjct: 388 ADITILASRDAVFLTGGPDYAVPLGRRDGVSFSTVGTQKLPSPINNTTATLKAFADRNFD 567
Query: 199 DHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKV 258
D+V L GAHT G+ C F RL DP +++ A CP N
Sbjct: 568 ATDVVALSGAHTFGRAHCGTFFNRL------SPLDPNMDKTLAKNLTATCPAQ-NSTNTA 726
Query: 259 SLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEF 318
+LD +P FD ++ ++ + GV SDQ L D T+ +V ++A N F +F
Sbjct: 727 NLDIRTPNVFDNKYYLDLMNRQGVFTSDQDLLSDKRTKGLVNAFAVN-----QTLFFEKF 891
Query: 319 PKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
A+IKLS ++V T +GEIR C+ N
Sbjct: 892 VDAVIKLSQLDVLTGNQGEIRGRCNVVN 975
>TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%)
Length = 645
Score = 192 bits (489), Expect = 5e-50
Identities = 103/209 (49%), Positives = 144/209 (68%), Gaps = 8/209 (3%)
Frame = +2
Query: 28 MLLGSLMILMISFIIAQ---AQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRL 84
M +I+++ F++ Q ++L+ G+YS SC AE V+ + K+P IA L+R+
Sbjct: 17 MKFNCAIIVLVIFLLNQNAYSELEVGYYSYSCGMAEFIVKDEVRRSVTKNPGIAAGLVRM 196
Query: 85 HFHDCFVQGCDGSVLI---VGSSAERNALANL-SLRGFEVIEDAKSQLEAICPGVVSCAD 140
HFHDCF++GCD SVL+ ++AE+++ AN SLRGFEVI+ AK++LEA+C GVVSCAD
Sbjct: 197 HFHDCFIRGCDASVLLDSTPSNTAEKDSPANKPSLRGFEVIDSAKAKLEAVCKGVVSCAD 376
Query: 141 ILAVAARDAVYLSNGPSWSVPTGRRDGRIS-SSAQASNLPSPLDPVSVQRQKFAAKGLDD 199
I+A AARD+V L+ G + VP GRRDGRIS +S ++LP P V+ Q FA KGL
Sbjct: 377 IIAFAARDSVELAGGVGYDVPAGRRDGRISLASDTRTDLPPPTFNVNQLTQLFAKKGLTQ 556
Query: 200 HDLVTLVGAHTIGQTECRFFSYRLYNFTT 228
++VTL GAHTIG++ C FS RLYNF++
Sbjct: 557 EEMVTLSGAHTIGRSHCAAFSSRLYNFSS 643
>TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precursor (Atperox
P10) (ATP5a) , partial (42%)
Length = 753
Score = 189 bits (481), Expect = 4e-49
Identities = 98/188 (52%), Positives = 128/188 (67%), Gaps = 4/188 (2%)
Frame = +3
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
L ++++S +++ +QL FY ++CP VR + S + D IA SL+RLHFHDCFV
Sbjct: 186 LWLVLLSPLVS-SQLYYNFYDTTCPNLTRIVRYNLLSAMSNDTRIAASLLRLHFHDCFVN 362
Query: 93 GCDGSVLIVGSSA---ERNALANL-SLRGFEVIEDAKSQLEAICPGVVSCADILAVAARD 148
GCDGSVL+ +S E+NAL N S+RGFEVI+ KS LE CP VSCADIL +AAR+
Sbjct: 363 GCDGSVLLDDTSTQKGEKNALPNKNSIRGFEVIDTIKSALEKACPSTVSCADILTLAARE 542
Query: 149 AVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGA 208
AVYLS GP WSVP GRRDG +S ++A+NLPSP +P+ KF +KGL+ D+ L GA
Sbjct: 543 AVYLSRGPFWSVPLGRRDGTTASESEANNLPSPFEPLENITAKFISKGLEKKDVAVLSGA 722
Query: 209 HTIGQTEC 216
HT G +C
Sbjct: 723 HTFGFAQC 746
>BP072224
Length = 453
Score = 187 bits (476), Expect = 2e-48
Identities = 93/95 (97%), Positives = 94/95 (98%)
Frame = -3
Query: 252 GNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLG 311
GNGLRKVSLDSGSPA+FDVSFFKNVRDGNGVLESD RLWGDSATQSVVQSYAGNIRGLLG
Sbjct: 451 GNGLRKVSLDSGSPAQFDVSFFKNVRDGNGVLESDPRLWGDSATQSVVQSYAGNIRGLLG 272
Query: 312 LKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
LKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN
Sbjct: 271 LKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 167
>TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase
precursor , partial (66%)
Length = 738
Score = 179 bits (455), Expect = 5e-46
Identities = 99/227 (43%), Positives = 135/227 (58%), Gaps = 6/227 (2%)
Frame = +1
Query: 34 MILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQG 93
++L + A AQL FY SCP + VR+ + K+ + S++RL FHDCFV G
Sbjct: 61 ILLSLLACSANAQLTPTFYDRSCPKLQTIVRNAMVQTIKKEARMGASILRLFFHDCFVNG 240
Query: 94 CDGSVLI--VGSS--AERNALANL-SLRGFEVIEDAKSQLEAICPGVVSCADILAVAARD 148
CDGS+L+ +G++ E+NA N S RGFEVI+ K+ +EA C VSCADILA+A RD
Sbjct: 241 CDGSILLDDIGTTFVGEKNAAPNKNSARGFEVIDTIKTNVEASCNNTVSCADILALATRD 420
Query: 149 AVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVG 207
+ L GP+W VP GRRD R +S ++A + +PSP +S F+AKGL DL L G
Sbjct: 421 GINLLGGPTWQVPLGRRDARTASQSKANTEIPSPSSDLSTLISMFSAKGLSARDLTVLSG 600
Query: 208 AHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNG 254
HTIGQ EC+FF R+ N++ I+ A A + CP +G G
Sbjct: 601 GHTIGQAECQFFRSRV-------NNETNIDAAFAASRKTNCPASGGG 720
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 168 bits (425), Expect = 1e-42
Identities = 113/345 (32%), Positives = 164/345 (46%), Gaps = 7/345 (2%)
Frame = +2
Query: 9 KGKREIPKIKYKVSSQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIE 68
K K+ PK S + L L + L FY +CP AE + ++
Sbjct: 38 KRKKMAPKAFIIFLSLLSLSALSLLSPSSAQEDVQDTGLAMNFYKETCPQAEDIITEQVK 217
Query: 69 SYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSS---AERNALANLSLRGFEVIEDAK 125
+ + A S +R FHDC VQ CD S+L+ + +E+ + LR F IE K
Sbjct: 218 LLYKRHKNTAFSWLRNIFHDCAVQSCDASLLLDSTRKTLSEKETDRSFGLRNFRYIETIK 397
Query: 126 SQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASN-LPSPLDP 184
LE CPGVVSC+DIL ++ARD + GP + TGRRDGR S + LP +
Sbjct: 398 EALERECPGVVSCSDILVLSARDGIAALGGPHIPLRTGRRDGRRSRADVVEQFLPDHNES 577
Query: 185 VSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQF 244
+S KF A G+D +V L+GAH++G+T C +RLY DP +N +
Sbjct: 578 ISAVLDKFGAMGIDTPGVVALLGAHSVGRTHCVKLVHRLY-----PEVDPALNPDHIPHM 742
Query: 245 QALCP---KNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQS 301
CP + ++ V D G+P D ++++N+ D G+L D +L D T+ V+
Sbjct: 743 LKKCPDAIPDPKAVQYVRNDRGTPMILDNNYYRNILDNKGLLLVDHQLANDKRTKPYVKK 922
Query: 302 YAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
A + F EF +A+ LS T T+GEIRK C+ N
Sbjct: 923 MAKSQD-----YFFKEFSRAITLLSENNPLTGTKGEIRKQCNVAN 1042
>TC17595 similar to UP|Q9XFI7 (Q9XFI7) Peroxidase (Fragment), partial (42%)
Length = 657
Score = 159 bits (401), Expect = 9e-40
Identities = 76/148 (51%), Positives = 109/148 (73%), Gaps = 3/148 (2%)
Frame = +1
Query: 27 AMLLGSLMILMISFIIAQA--QLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRL 84
A+++ +++L+ SF+I + QL+ GFYS++CP AE+ V++ + DP +A L+RL
Sbjct: 121 AVMVLFVLVLLFSFLIVSSDGQLQVGFYSNTCPHAESIVQAVVRGAVASDPNMAAVLLRL 300
Query: 85 HFHDCFVQGCDGSVLIV-GSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILA 143
HFHDCFV+GCDGS+LI G +E+ A + +RGFEVIE AK+Q EA CP VVSCADI+A
Sbjct: 301 HFHDCFVEGCDGSILIENGDQSEKLAFGHQGVRGFEVIERAKAQSEASCPDVVSCADIVA 480
Query: 144 VAARDAVYLSNGPSWSVPTGRRDGRISS 171
+AA D++ + NGP + VPTGRRDG +S+
Sbjct: 481 LAATDSIVMGNGPEYQVPTGRRDGSVSN 564
>TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
partial (47%)
Length = 604
Score = 157 bits (396), Expect = 3e-39
Identities = 79/168 (47%), Positives = 117/168 (69%), Gaps = 5/168 (2%)
Frame = +1
Query: 19 YKVSSQMEAMLLGSLMILMISFIIAQ--AQLKTGFYSSSCPTAEATVRSTIESYFNKDPT 76
Y+V +M ++I+ + +IA AQL+ GFY+ +CP AE + + + + P+
Sbjct: 73 YQVHMEMGGQSCFKVLIVCLLALIASNHAQLEQGFYTKTCPKAEKIILDFVHEHIHNAPS 252
Query: 77 IAPSLIRLHFHDCFVQGCDGSVLIVGSS--AERNALANLSLRGFEVIEDAKSQLEAICPG 134
+A +LIR++FHDCFV+GCD S+L+ +S AER+A NL++RGF+ I+ KS +EA CPG
Sbjct: 253 LAAALIRMNFHDCFVRGCDASILLNSTSKQAERDAPPNLTVRGFDFIDRIKSLVEAQCPG 432
Query: 135 VVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSP 181
VVSCADI+A+AARD++ + GP W VPTGRRDG IS+ +A S +P+P
Sbjct: 433 VVSCADIIALAARDSIVATGGPFWKVPTGRRDGVISNLVEARSQIPAP 576
>AV426241
Length = 429
Score = 153 bits (387), Expect = 4e-38
Identities = 76/140 (54%), Positives = 99/140 (70%)
Frame = +3
Query: 26 EAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLH 85
++ LL L++L + GFYS SCP+ E+ V+ST+ S+ D A L+RLH
Sbjct: 12 KSFLLVFLIVLTLQAFAVHGT-SVGFYSKSCPSIESIVKSTVASHVKTDFEYAAGLLRLH 188
Query: 86 FHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVA 145
F DCFV+GCD S+LI G+ E+ A N SL+G+EVI++AK++LEA CPGVVSCADILA+A
Sbjct: 189 FRDCFVRGCDASILIAGNGTEKQAPPNRSLKGYEVIDEAKAKLEAQCPGVVSCADILALA 368
Query: 146 ARDAVYLSNGPSWSVPTGRR 165
ARD+V LS G SW VPTGRR
Sbjct: 369 ARDSVVLSGGLSWQVPTGRR 428
>TC18103 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (37%)
Length = 546
Score = 141 bits (355), Expect(2) = 3e-36
Identities = 73/141 (51%), Positives = 96/141 (67%), Gaps = 4/141 (2%)
Frame = +1
Query: 11 KREIPKIKYKVSSQMEAMLLG-SLMILMISFIIAQAQ---LKTGFYSSSCPTAEATVRST 66
+R + KI Y ME L + ++L ++ I+ + GFY +CP AE+ VRS
Sbjct: 73 RRYLCKITYL*LINMERSLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSA 252
Query: 67 IESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKS 126
+ES+ D T+A L+R+HFHDCFVQGCD SVLI G+ ER A NL LRG+EVI+DAK+
Sbjct: 253 VESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGTERTAPPNLGLRGYEVIDDAKA 432
Query: 127 QLEAICPGVVSCADILAVAAR 147
++EA CPGVVSCADILA+AAR
Sbjct: 433 KVEAACPGVVSCADILALAAR 495
Score = 26.9 bits (58), Expect(2) = 3e-36
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = +2
Query: 148 DAVYLSNGPSWSVPTG 163
D+V LS G SW VPTG
Sbjct: 497 DSVVLSGGLSWQVPTG 544
>TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%)
Length = 559
Score = 140 bits (352), Expect = 4e-34
Identities = 80/181 (44%), Positives = 111/181 (61%), Gaps = 8/181 (4%)
Frame = +3
Query: 14 IPKIKYKVSSQMEAMLLGSLMILMISFIIAQ--AQLKTGFYSSSCPTAEATVRSTIESYF 71
I K+ SS A + L + M+ I + AQL FY CP+ V+S + S
Sbjct: 6 ISKMALSSSSSSSAANIFVLSLFMLFLIGSSNSAQLSENFYVKKCPSVFNAVKSVVHSAV 185
Query: 72 NKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSA---ERNALANL-SLRGFEVIEDAKSQ 127
K+ + SL+RL FHDCFV GCDGSVL+ +S+ E+ A N SLRGF+VI+ KS+
Sbjct: 186 AKEARMGGSLLRLFFHDCFVNGCDGSVLLDDTSSFKGEKTAPPNSNSLRGFDVIDAIKSK 365
Query: 128 LEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRISS--SAQASNLPSPLDPV 185
+EA+CPGVVSCAD++A+AARD+V + GP W V GRRD + +S +A + +PSP +
Sbjct: 366 VEAVCPGVVSCADVVAIAARDSVAILGGPYWKVKLGRRDSKTASFNAANSGVIPSPFSSL 545
Query: 186 S 186
S
Sbjct: 546 S 548
>TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (49%)
Length = 581
Score = 128 bits (322), Expect = 1e-30
Identities = 78/187 (41%), Positives = 116/187 (61%), Gaps = 10/187 (5%)
Frame = +2
Query: 20 KVSSQMEAMLLGSLMILMISFIIAQ--AQLKTGFYSSSCPTAEATVRSTIESYFNKDPTI 77
K++S ++ L+ +++ + +F+I A L G+Y CP A ++S ++ N++ I
Sbjct: 23 KMNSHRQSFLV-LVLVTLATFLIPSTLAGLTPGYYDKVCPQALPIIKSVVQKAINREQRI 199
Query: 78 APSLIRLHFHDCFVQGCDGSVLIVGSSA---ERNALANL-SLRGFEVIEDAKSQLEAICP 133
SL+RLHFHDCFV GCDGS+L+ +S+ E+ AL NL S+RGFEV+++ K+ ++ C
Sbjct: 200 GASLLRLHFHDCFVNGCDGSLLLDDTSSFVGEKTALPNLNSVRGFEVVDEIKAAVDRACK 379
Query: 134 -GVVSCADILAVAARDAVYLSNGPS--WSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQR 189
VVSCADILA+AARD+V + G + V GRRD R +S A A SNLP P +
Sbjct: 380 RPVVSCADILAIAARDSVAILGGKQYWYQVRLGRRDARSASQAAANSNLPPPFLNFTQLL 559
Query: 190 QKFAAKG 196
F A+G
Sbjct: 560 PAFQAQG 580
>AV412471
Length = 430
Score = 127 bits (320), Expect = 2e-30
Identities = 58/122 (47%), Positives = 86/122 (69%), Gaps = 3/122 (2%)
Frame = +2
Query: 35 ILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGC 94
+++ F L +Y SCP + V++ + + DPT+A LIR+HFHDCF++GC
Sbjct: 62 VIVSGFTFGVDDLNMNYYLFSCPFVDPIVKNKVSTALQNDPTLAAGLIRMHFHDCFIEGC 241
Query: 95 DGSVLIVGS---SAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVY 151
DGSVL+ + +AE+++ ANLSLRG+E+I+D K +LE CPGVVSCADI+A+A+RDAV+
Sbjct: 242 DGSVLLDSTKDNTAEKDSPANLSLRGYEIIDDIKEELEKRCPGVVSCADIVAMASRDAVF 421
Query: 152 LS 153
+
Sbjct: 422 FA 427
>AV414604
Length = 427
Score = 125 bits (315), Expect = 8e-30
Identities = 62/130 (47%), Positives = 88/130 (67%), Gaps = 3/130 (2%)
Frame = +2
Query: 24 QMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIR 83
++ + LL L + + L+ FY SC AE V++TI+ + + P + L+R
Sbjct: 17 KINSPLLACLAMFCFLGVCQGGSLRKQFYRKSCSQAEQIVKTTIQQHVSSRPELPAKLLR 196
Query: 84 LHFHDCFVQGCDGSVLI---VGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCAD 140
+HFHDCFV+GCDGSVL+ G++AE++A+ NLSL GF+VI++ K LEA CP +VSCAD
Sbjct: 197 MHFHDCFVRGCDGSVLLNSTAGNTAEKDAIPNLSLSGFDVIDEIKEALEAKCPKIVSCAD 376
Query: 141 ILAVAARDAV 150
ILA+AARDAV
Sbjct: 377 ILALAARDAV 406
>BP051104
Length = 566
Score = 124 bits (310), Expect = 3e-29
Identities = 75/168 (44%), Positives = 96/168 (56%), Gaps = 5/168 (2%)
Frame = +2
Query: 91 VQGCDGSVLIVGSS---AERNALAN-LSLRGFEVIEDAKSQLEAICPGVVSCADILAVAA 146
V GCD S+L +S E+ A AN S RGF VI+ K+ LE CPGVVSCAD+LA+AA
Sbjct: 71 VNGCDASILXDDTSNFIGEQTAAANNRSARGFNVIDGIKANLEKQCPGVVSCADVLALAA 250
Query: 147 RDAVYLSNGPSWSVPTGRRDGRISSSAQASN-LPSPLDPVSVQRQKFAAKGLDDHDLVTL 205
RD+V GPSW V GRRD +S A+N +P P +S FA +GL DLV L
Sbjct: 251 RDSVVQLGGPSWEVGLGRRDSTTASRGTANNTIPGPFLSLSGLITNFANQGLSVTDLVAL 430
Query: 206 VGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGN 253
GAHTIG +C+ F +YN D I+ + + CP++GN
Sbjct: 431 SGAHTIGLAQCKNFRAHIYN-------DSNIDASYAKFLKX*CPRSGN 553
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,169,208
Number of Sequences: 28460
Number of extensions: 63662
Number of successful extensions: 420
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 362
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 365
length of query: 346
length of database: 4,897,600
effective HSP length: 91
effective length of query: 255
effective length of database: 2,307,740
effective search space: 588473700
effective search space used: 588473700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0252a.1


