

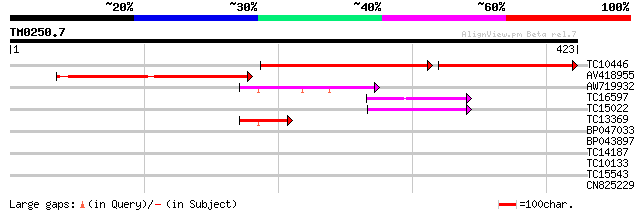
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0250.7
(423 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10446 similar to UP|Q8GTR1 (Q8GTR1) Geranylgeranyl diphosphate... 235 e-109
AV418955 205 1e-53
AW719932 53 8e-08
TC16597 similar to UP|Q9SST9 (Q9SST9) Geranylgeranyl pyrophospha... 46 1e-05
TC15022 similar to UP|Q94ID7 (Q94ID7) Geranylgeranyl diphosphate... 45 2e-05
TC13369 similar to UP|Q9FPK0 (Q9FPK0) AT4g38460, partial (35%) 42 2e-04
BP047033 37 0.006
BP043897 28 3.7
TC14187 weakly similar to UP|Q9ZTM8 (Q9ZTM8) PGPS/D12, partial (... 28 3.7
TC10133 similar to UP|Q9LSS5 (Q9LSS5) Myosin heavy chain-like, p... 28 3.7
TC15543 similar to UP|Q84K86 (Q84K86) 4-coumarate:CoA ligase-lik... 27 8.3
CN825229 27 8.3
>TC10446 similar to UP|Q8GTR1 (Q8GTR1) Geranylgeranyl diphosphate synthase,
partial (54%)
Length = 999
Score = 235 bits (599), Expect(2) = e-109
Identities = 119/128 (92%), Positives = 125/128 (96%)
Frame = +1
Query: 188 ADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQ 247
ADTRRGIGSLNFVM NKL +L GDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQ
Sbjct: 1 ADTRRGIGSLNFVMANKLPLLPGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQ 180
Query: 248 MSTTSDQRCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQL 307
M+TTSDQRCSMEYYM+KTYYKTASLISNSCKA+AILAGQTA+VAMLAF+YGKNLGLAFQL
Sbjct: 181 MTTTSDQRCSMEYYMEKTYYKTASLISNSCKALAILAGQTAEVAMLAFEYGKNLGLAFQL 360
Query: 308 IDDVLDFT 315
IDDVLDFT
Sbjct: 361 IDDVLDFT 384
Score = 177 bits (448), Expect(2) = e-109
Identities = 88/103 (85%), Positives = 98/103 (94%)
Frame = +3
Query: 321 LGKGSLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRE 380
LGKGSLSDIRHGIVTAPILFAMEEFPQLR +V++GF+NPENVD+ALEYLGKSRGIQRT+E
Sbjct: 402 LGKGSLSDIRHGIVTAPILFAMEEFPQLRAIVEDGFENPENVDLALEYLGKSRGIQRTKE 581
Query: 381 LAIKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVITRTK 423
LA+KHA+LAAAAIDSLPESDD EVR SR+AL+DLT VITRTK
Sbjct: 582 LAMKHANLAAAAIDSLPESDDEEVRRSRKALIDLTHIVITRTK 710
>AV418955
Length = 413
Score = 205 bits (522), Expect = 1e-53
Identities = 112/146 (76%), Positives = 119/146 (80%)
Frame = +3
Query: 36 QSHHYHTPTGSTAKVMRSLAFSKGLPALHSSRYQIHDQSSSISEEELDPFSLVADELSFI 95
QSH T ST +V+RSL FSKG PAL+SSR+QIH QSS I E+E DPFSLVADELS +
Sbjct: 3 QSH-----TDSTQQVIRSLIFSKGFPALYSSRHQIHHQSSPIVEDEHDPFSLVADELSIL 167
Query: 96 ANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKPPPSNDLGGT 155
N LR MV PKLAS AEYFFKMGVEGKRFRPTVLLLMSTALNLPIPK PP +LGGT
Sbjct: 168 GNKLREMV----PKLASAAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKEPPPIELGGT 335
Query: 156 FTADLRSRQQRIAEITEMIHVASLLH 181
T DLRSRQQRIAEITEMIHVASLLH
Sbjct: 336 MTTDLRSRQQRIAEITEMIHVASLLH 413
>AW719932
Length = 402
Score = 53.1 bits (126), Expect = 8e-08
Identities = 41/114 (35%), Positives = 59/114 (50%), Gaps = 9/114 (7%)
Frame = +1
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRA-----CVALASL 224
E+IH SL+HDD+ +D+ D RRG + + G AVLAGD LL+ A C +A
Sbjct: 55 EIIHTMSLIHDDLPCMDNDDLRRGRPTNHKAFGENFAVLAGDALLAFAFEHLTCSKVAPG 234
Query: 225 KNTEVVSLLAKVV--EHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNS 276
+ V+ LAK V E LV G+T+ + + +E +KTA L+ S
Sbjct: 235 RVVRAVAELAKSVGGEGLVAGQTVDIDSEGVSDVGLERLEYIHVHKTAVLLEAS 396
>TC16597 similar to UP|Q9SST9 (Q9SST9) Geranylgeranyl pyrophosphate
synthase, partial (40%)
Length = 564
Score = 45.8 bits (107), Expect = 1e-05
Identities = 30/79 (37%), Positives = 46/79 (57%), Gaps = 1/79 (1%)
Frame = +3
Query: 267 YKTASLISNSCKAIAILAGQT-ADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGS 325
+KTA+L+ + AIL G + A+V L + +GL FQ++DD+LD T +S LGK +
Sbjct: 69 HKTAALLEGAVVLGAILGGGSDAEVERLR-KFASCIGLLFQVVDDILDVTKSSQELGKTA 245
Query: 326 LSDIRHGIVTAPILFAMEE 344
D+ VT P L +E+
Sbjct: 246 GKDLVADKVTYPKLLGIEK 302
>TC15022 similar to UP|Q94ID7 (Q94ID7) Geranylgeranyl diphosphate synthase
, partial (31%)
Length = 553
Score = 45.4 bits (106), Expect = 2e-05
Identities = 27/77 (35%), Positives = 42/77 (54%)
Frame = +1
Query: 268 KTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLS 327
KTA L+ S AI+ G + + + + +GL FQ++DD+LD T +S LGK +
Sbjct: 1 KTAVLLEASVVLGAIMGGGSDEEVERLRKFARCIGLLFQVVDDILDVTKSSEELGKTAGK 180
Query: 328 DIRHGIVTAPILFAMEE 344
D+ VT P L +E+
Sbjct: 181 DLVADKVTYPKLLGIEK 231
>TC13369 similar to UP|Q9FPK0 (Q9FPK0) AT4g38460, partial (35%)
Length = 559
Score = 41.6 bits (96), Expect = 2e-04
Identities = 20/42 (47%), Positives = 30/42 (70%), Gaps = 2/42 (4%)
Frame = +1
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGD 211
EM+H AS +HDD+ +DD+ +RRG S + + G +A+LAGD
Sbjct: 433 EMVHAASWIHDDLPCMDDSPSRRGQPSNHAIYGVDMAILAGD 558
>BP047033
Length = 379
Score = 37.0 bits (84), Expect = 0.006
Identities = 19/45 (42%), Positives = 26/45 (57%)
Frame = -1
Query: 302 GLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEEFP 346
GL FQ++DD+LD T +S LG+ + D+ VT P F E P
Sbjct: 379 GLLFQVVDDILDVTKSSPGLGETAGKDLVADKVTYPHAFGDREVP 245
>BP043897
Length = 472
Score = 27.7 bits (60), Expect = 3.7
Identities = 23/87 (26%), Positives = 34/87 (38%), Gaps = 9/87 (10%)
Frame = -2
Query: 142 PIPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADT---------RR 192
P +P S +GG R R + + V +++ DD+L DT R+
Sbjct: 375 PTARPRRSASVGGWPPPSWRERSSGA*GSSSGVSVVAVVRDDILKARDTKSAFA*LKLRK 196
Query: 193 GIGSLNFVMGNKLAVLAGDFLLSRACV 219
G G V KL + F +S CV
Sbjct: 195 GEGFRKHVWS*KLEIRVIQFQISINCV 115
>TC14187 weakly similar to UP|Q9ZTM8 (Q9ZTM8) PGPS/D12, partial (66%)
Length = 1030
Score = 27.7 bits (60), Expect = 3.7
Identities = 13/51 (25%), Positives = 24/51 (46%)
Frame = -2
Query: 229 VVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKA 279
++S + HL E +TTS+Q C + ++ SL++N C +
Sbjct: 483 IISFQMVLRSHLAPVEA*VQATTSNQTCDQSVQSPTSTTRSGSLVNNLCNS 331
>TC10133 similar to UP|Q9LSS5 (Q9LSS5) Myosin heavy chain-like, partial (34%)
Length = 1827
Score = 27.7 bits (60), Expect = 3.7
Identities = 16/63 (25%), Positives = 29/63 (45%)
Frame = -2
Query: 213 LLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASL 272
LLS +C+A+ S + LA + L+ +S +SD C++ + + A+
Sbjct: 1313 LLSTSCIAIPSFMRASCSAALAASISSLLFLSNFSISFSSDMLCNVVSLSIISAFNAATQ 1134
Query: 273 ISN 275
SN
Sbjct: 1133 CSN 1125
>TC15543 similar to UP|Q84K86 (Q84K86) 4-coumarate:CoA ligase-like, partial
(21%)
Length = 579
Score = 26.6 bits (57), Expect = 8.3
Identities = 17/54 (31%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Frame = +1
Query: 357 DNPENVDIA--LEYLGKSRGIQ----RTRELAIKHADLAAAAIDSLPESDDGEV 404
D+ ENV I ++ L K +G Q + + H + AA+ SLP+ + GE+
Sbjct: 4 DDEENVFIVDRIKELIKYKGFQVAPAELEAILLSHPSIEDAAVVSLPDEEAGEI 165
>CN825229
Length = 641
Score = 26.6 bits (57), Expect = 8.3
Identities = 13/38 (34%), Positives = 23/38 (60%)
Frame = +3
Query: 383 IKHADLAAAAIDSLPESDDGEVRLSRRALVDLTQRVIT 420
I+H D + + LP +D + RL+ + L+D++ VIT
Sbjct: 3 IRHED--SILLRELPPVEDADSRLALKRLIDISMGVIT 110
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,943,653
Number of Sequences: 28460
Number of extensions: 77181
Number of successful extensions: 587
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 584
length of query: 423
length of database: 4,897,600
effective HSP length: 93
effective length of query: 330
effective length of database: 2,250,820
effective search space: 742770600
effective search space used: 742770600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0250.7


