

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0248.4
(573 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
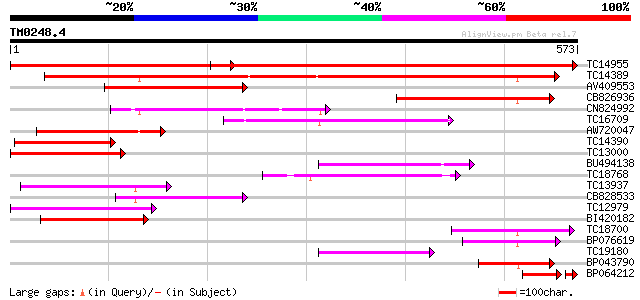
Score E
Sequences producing significant alignments: (bits) Value
TC14955 UP|O22305 (O22305) Peptide transporter, complete 742 0.0
TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 428 e-120
AV409553 183 8e-47
CB826936 139 1e-33
CN824992 130 6e-31
TC16709 weakly similar to PIR|T47573|T47573 peptide transport-li... 127 5e-30
AW720047 122 1e-28
TC14390 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 118 3e-27
TC13000 similar to PIR|T47573|T47573 peptide transport-like prot... 110 5e-25
BU494138 110 6e-25
TC18768 104 4e-23
TC13937 weakly similar to UP|Q9LVE0 (Q9LVE0) Nitrate transporter... 98 3e-21
CB828533 97 7e-21
TC12979 similar to UP|Q9LQL2 (Q9LQL2) F5D14.23 protein, partial ... 96 2e-20
BI420182 93 1e-19
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 84 6e-17
BP076619 82 2e-16
TC19180 similar to PIR|T10255|T10255 nitrite transport protein, ... 78 4e-15
BP043790 78 4e-15
BP064212 70 5e-15
>TC14955 UP|O22305 (O22305) Peptide transporter, complete
Length = 2300
Score = 742 bits (1915), Expect = 0.0
Identities = 365/370 (98%), Positives = 366/370 (98%)
Frame = +1
Query: 204 QEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQL 263
+ K GLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQL
Sbjct: 646 KRKMDXGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQL 825
Query: 264 PCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPPCTVTQVEGTKLV 323
PCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPPCTVTQVEGTKLV
Sbjct: 826 PCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPPCTVTQVEGTKLV 1005
Query: 324 LGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPI 383
LGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPI
Sbjct: 1006 LGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPI 1185
Query: 384 YDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEE 443
YDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEE
Sbjct: 1186 YDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEE 1365
Query: 444 TVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTF 503
TVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTF
Sbjct: 1366 TVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTF 1545
Query: 504 LVTMIDKLNWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTSTTEVHDI 563
LVTMIDKLNWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTSTTEVHDI
Sbjct: 1546 LVTMIDKLNWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTSTTEVHDI 1725
Query: 564 EMSAEKTVKY 573
EMSAEKTVKY
Sbjct: 1726 EMSAEKTVKY 1755
Score = 425 bits (1092), Expect = e-119
Identities = 212/228 (92%), Positives = 218/228 (94%)
Frame = +3
Query: 1 MEGKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLN 60
MEGKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLN
Sbjct: 36 MEGKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLN 215
Query: 61 KDVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKS 120
KDVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKS
Sbjct: 216 KDVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKS 395
Query: 121 LRPACENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVS 180
LRPACENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVS
Sbjct: 396 LRPACENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVS 575
Query: 181 FFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFF 228
FFNWWAFNGACGSLMATLFVVYIQEKNG G + L I + S+ +++
Sbjct: 576 FFNWWAFNGACGSLMATLFVVYIQEKNGXG--FGLQFICYWFSTFLYY 713
>TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(72%)
Length = 1990
Score = 428 bits (1100), Expect = e-120
Identities = 216/532 (40%), Positives = 324/532 (60%), Gaps = 12/532 (2%)
Frame = +3
Query: 36 YRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILGAYIADTYTGRF 95
Y + ER AYYG+ +NLV F+ ++L++ V+S + +NW+G+ TP++GAYIAD Y GR+
Sbjct: 27 YELFERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRY 206
Query: 96 WTITFSLLIYAIGLVLLVLTTTLKSLRPA-CENGI----CREASNLQVALFYTSLYTIAV 150
WT + IY +G+ LL LT +L +LRP C G+ C +AS +FY +LY IA+
Sbjct: 207 WTFIIASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIAL 386
Query: 151 GSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKNGWG 210
G+G KPN+ST GADQFD++ +E K+SFFNWW F+ G L +T +VYIQ+ W
Sbjct: 387 GAGGTKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWS 566
Query: 211 LAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSEL 270
L Y L +G S ++F G+P YRHK SP ++V + A K +P +P +L
Sbjct: 567 LGYGLPTVGLAFSILVFLVGTPFYRHKLPSG-SPLTRMLQVFVAAGIKWKAHVPHDPKQL 743
Query: 271 HEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPPCTVTQVEGTKLVLGMFQIW 330
HE + Y ++ +I HTS FLD+AA++ T CTVTQVE TK + M I
Sbjct: 744 HELSMEEYANNSRNRIDHTSTLRFLDKAAVKTGKTSPWRL-CTVTQVEETKQMTKMVPIL 920
Query: 331 LLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIP 390
+ LIP T+F++QGTT+DR++GP F +P A L ++++ L +PIYD F+P
Sbjct: 921 ITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVP 1100
Query: 391 FMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIF 450
+RR T N RGI +LQR+G G+ + V+ M + + E +R+ V ++ H++G + +P+SIF
Sbjct: 1101VIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIF 1280
Query: 451 WLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDK 510
L+PQ + GV+ F ++FFY Q+PE MK LG + T+ VA G ++++FL++ +
Sbjct: 1281ILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVAD 1460
Query: 511 L-------NWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENT 555
+ WI +NLN S LDYYYAF+ ++S LNF FL + + Y + T
Sbjct: 1461ITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVT 1616
>AV409553
Length = 433
Score = 183 bits (464), Expect = 8e-47
Identities = 81/144 (56%), Positives = 105/144 (72%)
Frame = +2
Query: 97 TITFSLLIYAIGLVLLVLTTTLKSLRPACENGICREASNLQVALFYTSLYTIAVGSGAVK 156
T T S ++Y +G++LL + +LKSL+P C NGIC +AS Q+ FYT+LYT A+G+G K
Sbjct: 2 TFTLSSIVYVMGMILLTVAVSLKSLKPTCTNGICNKASTSQIVFFYTALYTTAIGAGGTK 181
Query: 157 PNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLS 216
PN+STFGADQFDDF EK+ K SFFNWW F G+L+ATL +VYIQE GWGL Y +
Sbjct: 182 PNISTFGADQFDDFNPHEKQTKASFFNWWMFTSFLGALIATLGLVYIQENLGWGLGYGIP 361
Query: 217 AIGFLLSSIIFFWGSPVYRHKSRQ 240
G LLS +IF+ G+P+YRHK R+
Sbjct: 362 TSGLLLSLVIFYVGTPMYRHKVRK 433
>CB826936
Length = 596
Score = 139 bits (351), Expect = 1e-33
Identities = 68/166 (40%), Positives = 107/166 (63%), Gaps = 7/166 (4%)
Frame = +3
Query: 392 MRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFW 451
MR+ T N RGI +LQR+G G+ + +I M + E +R+SV ++++++G + +P++IF
Sbjct: 6 MRQYTKNPRGIAMLQRLGAGLVMYIITMVTAWLSERKRLSVAREYNLLGQHDKIPLTIFI 185
Query: 452 LLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL 511
LLPQ + GV+N F+ ML+FFYDQ+PE MK LG + T+ A G + ++FL++ + L
Sbjct: 186 LLPQFALTGVANNFVDIAMLDFFYDQAPEGMKSLGISYSTTSTALGCFFSSFLLSTVADL 365
Query: 512 -------NWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIY 550
W+ +NLN SHLDYYYAF+ ++S NF FL + + Y
Sbjct: 366 TKKHGHKGWVLDNLNVSHLDYYYAFMAILSVANFLCFLVAAKLFAY 503
>CN824992
Length = 708
Score = 130 bits (327), Expect = 6e-31
Identities = 85/235 (36%), Positives = 127/235 (53%), Gaps = 13/235 (5%)
Frame = +2
Query: 103 LIYAIGLVLLVLTTTLKSLRPACENGI------CREASNLQVALFYTSLYTIAVGSGAVK 156
+I+ +GL L LTT L L P NG C S+ Q ALFY S+Y IA+G+G +
Sbjct: 14 VIFVVGLASLSLTTYLFLLEP---NGCGDKELPCTSHSSYQTALFYVSIYLIALGNGGYQ 184
Query: 157 PNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLS 216
PN++TFGADQFD+ +E+ K+SFF+++ GSL + + Y ++ W L + S
Sbjct: 185 PNIATFGADQFDEEDPKEQHSKISFFSYFYLALNLGSLFSNTILNYFEDDGLWTLGFWAS 364
Query: 217 AIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQL-PCNPSELHEFQL 275
A L+ ++F G+P YR+ + +P F +V + A R K+++ P +LHE +
Sbjct: 365 AGSAFLALVLFLCGTPWYRY-FKPGGNPVPRFCQVFVAAMRKWKVKVSPGEEDQLHE--V 535
Query: 276 NYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPPC------TVTQVEGTKLVL 324
++G RK+ HT F FLD+AA+ S C TVTQVE K +L
Sbjct: 536 EECSTNGGRKMFHTEGFRFLDKAALITSQESKQENECSLWHLSTVTQVEEVKCIL 700
>TC16709 weakly similar to PIR|T47573|T47573 peptide transport-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(41%)
Length = 712
Score = 127 bits (319), Expect = 5e-30
Identities = 76/238 (31%), Positives = 129/238 (53%), Gaps = 6/238 (2%)
Frame = +1
Query: 217 AIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLN 276
A+ ++ + FF G+ +YR++ + SP +V + + R +Q P + S L+E
Sbjct: 4 AVAMAVAVVSFFSGTRLYRNQ-KPGGSPLTRMCQVVVASMRKCGVQAPDDKSLLYEIADT 180
Query: 277 YYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPP-----CTVTQVEGTKLVLGMFQIWL 331
G+RK+ HT+ SF D+AA+ ++D + CT TQVE K +L M +W
Sbjct: 181 ESAIKGSRKLDHTNELSFFDKAAVVVQSSDQKDSVDPWRLCTETQVEELKSILRMLPVWA 360
Query: 332 LMLIPTNCWALESTIFVRQGTTMDRTLG-PKFRLPAASLWCFIVLTTLICLPIYDHYFIP 390
+I + ST+FV QG TM+ +G F++P A+L F L+ + +P+YD +P
Sbjct: 361 TGIIFATVYGQMSTLFVLQGQTMNTHVGNSNFKIPPAALSIFDTLSVIFWVPVYDWIIVP 540
Query: 391 FMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMS 448
R+ +G+ G+ LQR+GIG+ I + AM +E R+ ++++H+ +E VPMS
Sbjct: 541 IARKFSGHKNGLTQLQRMGIGLFISIFAMVAAAILEVIRLRMVRRHNYYELKE-VPMS 711
>AW720047
Length = 419
Score = 122 bits (307), Expect = 1e-28
Identities = 58/131 (44%), Positives = 88/131 (66%), Gaps = 1/131 (0%)
Frame = +3
Query: 28 KACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILGAYI 87
KAC +IL ER AYYG+ NLV ++TT+LN+ VS+ + N + G LTP++G+++
Sbjct: 9 KACPFILGSFFCERLAYYGIATNLVIYLTTKLNEGTVSAAKNVNTFHGTCYLTPLIGSFL 188
Query: 88 ADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPA-CENGICREASNLQVALFYTSLY 146
AD Y GR+WTIT +Y IG+ +L+L+ T+ +L+PA C N +C A+ Q F+ LY
Sbjct: 189 ADAYWGRYWTITVFYAVYLIGMCILILSATIPALKPAECVNSVC-TATLAQSVXFFLGLY 365
Query: 147 TIAVGSGAVKP 157
+IA+G+G +KP
Sbjct: 366 SIALGTGGIKP 398
>TC14390 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(18%)
Length = 622
Score = 118 bits (295), Expect = 3e-27
Identities = 53/102 (51%), Positives = 73/102 (70%)
Frame = +1
Query: 6 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 65
YT DGTVDL GRPVL S TG KACT+I+ Y + ER AYYG+ +NLV F+ ++L++ V+
Sbjct: 82 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 261
Query: 66 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAI 107
S + +NW+G+ TP++GAYIAD Y GR+WT + IY +
Sbjct: 262 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLL 387
>TC13000 similar to PIR|T47573|T47573 peptide transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(21%)
Length = 446
Score = 110 bits (276), Expect = 5e-25
Identities = 52/116 (44%), Positives = 71/116 (60%)
Frame = +3
Query: 2 EGKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNK 61
E YT DGTVD G P TG KAC +IL ER AYYG+ NLV + QLN+
Sbjct: 99 EDDNYTKDGTVDYLGNPANKKQTGTWKACPFILGNECCERLAYYGMSTNLVLYFKKQLNQ 278
Query: 62 DVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTT 117
++ + +NWSG +TP++GA++AD+Y GR+WTI +IY IG+ LL L+ +
Sbjct: 279 HSATASKNVSNWSGTCYITPLIGAFLADSYLGRYWTIACFSIIYVIGMTLLTLSAS 446
>BU494138
Length = 518
Score = 110 bits (275), Expect = 6e-25
Identities = 60/157 (38%), Positives = 86/157 (54%)
Frame = +2
Query: 313 TVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCF 372
++ QVE K + +F IW ++ A + T V Q MDR +G KF++PA SL
Sbjct: 20 SIQQVEEIKCLARIFPIWAAGILGFTAMAQQGTFTVSQAMKMDRHIGSKFQIPAGSLGVI 199
Query: 373 IVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSV 432
+T + +P YD +F+P +RR T + GI LLQR+GIGM V++M V VE R V
Sbjct: 200 SFITIGLWVPFYDRFFVPAVRRITKHEGGITLLQRIGIGMVFSVLSMIVAGLVEKVRRGV 379
Query: 433 IKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATG 469
+ P PMS+ WL PQ +++G+ AF A G
Sbjct: 380 ANSN--PNPLGIAPMSVMWLAPQLVLMGLCEAFNAIG 484
>TC18768
Length = 738
Score = 104 bits (259), Expect = 4e-23
Identities = 71/205 (34%), Positives = 100/205 (48%), Gaps = 5/205 (2%)
Frame = +2
Query: 256 FRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRE----SNTDLSN-P 310
F RK P NP LH Q N HT F FLD+A IR SNT S+
Sbjct: 161 FFKRKHIYPSNPQMLHGDQNNVGQV-------HTDKFRFLDKACIRVEEAGSNTKKSSWR 319
Query: 311 PCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLW 370
C+V QVE K++L + I+ ++ A T V+QG+ MD L F +P ASL
Sbjct: 320 LCSVGQVEQVKILLSVIPIFSCTIVFNTILAQLQTFSVQQGSAMDTHLTKSFHIPPASLQ 499
Query: 371 CFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRM 430
+ +I +P+YD +F+PF RR TG+ GI L+R+G G+ + +M +E +R
Sbjct: 500 SIPYILLIIVVPLYDTFFVPFARRITGHESGISPLRRIGFGLFLATFSMVSAALLEKKRR 679
Query: 431 SVIKKHHIVGPEETVPMSIFWLLPQ 455
H+ +SIFW+ PQ
Sbjct: 680 DEALNHN-------KTLSIFWITPQ 733
>TC13937 weakly similar to UP|Q9LVE0 (Q9LVE0) Nitrate transporter
(AT3g21670/MIL23_23), partial (27%)
Length = 558
Score = 98.2 bits (243), Expect = 3e-21
Identities = 60/158 (37%), Positives = 83/158 (51%), Gaps = 6/158 (3%)
Frame = +2
Query: 12 VDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFN 71
VD GRP S TG A IL + ++ G+ N+V ++ LN S T
Sbjct: 83 VDYLGRPPDKSKTGGWTAAWLILGVELAQKIMVLGISMNIVTYLVGVLNLPTAYSATIVT 262
Query: 72 NWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLR-PAC----- 125
N G L +LG ++AD GR+ TI S I A+G+ LL TT+ S++ P C
Sbjct: 263 NCLGTMNLVSLLGGFLADVKLGRYLTIAISATISAVGMCLLTGATTIPSMKAPPCSSVQR 442
Query: 126 ENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFG 163
++ C EAS QVAL +LYTIAVG+G +K ++S FG
Sbjct: 443 QHHECIEASGKQVALLLVALYTIAVGAGGIKSSVSGFG 556
>CB828533
Length = 557
Score = 97.1 bits (240), Expect = 7e-21
Identities = 56/139 (40%), Positives = 83/139 (59%), Gaps = 6/139 (4%)
Frame = +1
Query: 108 GLVLLVLTTTLKSLR-PAC-----ENGICREASNLQVALFYTSLYTIAVGSGAVKPNMST 161
G+ LL TT+ S++ P C ++ C EAS QVAL +LYTIAVG+G +K ++S
Sbjct: 103 GMCLLTGATTIPSMKAPPCSSVQRQHHECIEASGKQVALLLVALYTIAVGAGGIKSSVSG 282
Query: 162 FGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFL 221
FG+DQFD E+++ V FFN + F + GSL + L +VY+Q+ G G + + A L
Sbjct: 283 FGSDQFDTTDPREEKKMVFFFNRFYFFVSTGSLFSVLVLVYVQDNIGRGWGFGIPAGIML 462
Query: 222 LSSIIFFWGSPVYRHKSRQ 240
+S +G P+YR+K Q
Sbjct: 463 VSLAFLLYGKPLYRYKRPQ 519
>TC12979 similar to UP|Q9LQL2 (Q9LQL2) F5D14.23 protein, partial (25%)
Length = 601
Score = 95.5 bits (236), Expect = 2e-20
Identities = 54/150 (36%), Positives = 88/150 (58%), Gaps = 3/150 (2%)
Frame = +3
Query: 2 EGKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNK 61
E + TLDG+VD GRP + + +G+ A T IL+ + L A++GVG NLV F+T L +
Sbjct: 150 EAEEVTLDGSVDWHGRPAIRAKSGRWVAGTIILLNQGLATLAFFGVGVNLVLFLTRVLGQ 329
Query: 62 DVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSL 121
D + + + W+G L ++GA+++D+Y GR+ T I+ +GLV L L++ L L
Sbjct: 330 DNADAANNVSKWTGTVYLFSLVGAFLSDSYWGRYKTCAIFQGIFVLGLVFLSLSSYLSLL 509
Query: 122 RP-ACENGI--CREASNLQVALFYTSLYTI 148
RP C + + C + S+L++ +F Y I
Sbjct: 510 RPKGCGSELLHCGKHSSLEMGMFSYQSYLI 599
>BI420182
Length = 506
Score = 93.2 bits (230), Expect = 1e-19
Identities = 41/111 (36%), Positives = 73/111 (64%), Gaps = 2/111 (1%)
Frame = +3
Query: 32 YILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILGAYIADTY 91
+I + E+ A G N+++++TTQL+ + + + N+ G A+LTP+LGA+I+D+Y
Sbjct: 171 FIFANEICEKLAVVGFSTNMISYLTTQLHMPLTKAANTLTNFGGTASLTPLLGAFISDSY 350
Query: 92 TGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENG--ICREASNLQVAL 140
G+FWTIT + ++Y IG+V L ++ L LRP G +C++A++ Q+A+
Sbjct: 351 AGKFWTITMASVLYQIGMVSLTISAVLPQLRPPPCRGEEVCKQATDGQLAV 503
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 84.0 bits (206), Expect = 6e-17
Identities = 38/131 (29%), Positives = 68/131 (51%), Gaps = 7/131 (5%)
Frame = +1
Query: 447 MSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVT 506
+S +WLL Q ++GV+ F G+LEF Y+++P+ MK +G+ G + T + +
Sbjct: 46 LSAYWLLIQYCLIGVAEVFCIVGLLEFLYEEAPDAMKSIGSAYAALAGGLGCFAATIINS 225
Query: 507 MIDKL-------NWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTSTTE 559
+I L +W+ N+N DY+Y L +S +NF +F++ + Y Y+ + E
Sbjct: 226 IIKSLTGKEGKESWLAQNINTGRFDYFYWILTALSLVNFCIFIYSAHRYKYRTQQVYEME 405
Query: 560 VHDIEMSAEKT 570
HD+ + T
Sbjct: 406 KHDVANNVSST 438
>BP076619
Length = 432
Score = 82.0 bits (201), Expect = 2e-16
Identities = 37/106 (34%), Positives = 59/106 (54%), Gaps = 7/106 (6%)
Frame = -3
Query: 458 ILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL------ 511
+ GVS F G+ EFFYDQ P E++ LG L S GS+++ FL+++I+ +
Sbjct: 427 LFGVSEVFTLVGLQEFFYDQVPNELRSLGLALYLSIFGVGSFLSGFLISVIETVTGKDGP 248
Query: 512 -NWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTS 556
W NNL+ +H+DY+Y L S + F +F+ + Y+Y + S
Sbjct: 247 DRWFANNLHKAHIDYFYWLLAGFSVVGFAMFMCFAKSYVYNHQGAS 110
>TC19180 similar to PIR|T10255|T10255 nitrite transport protein, chloroplast
- cucumber {Cucumis sativus;} , partial (12%)
Length = 408
Score = 77.8 bits (190), Expect = 4e-15
Identities = 44/117 (37%), Positives = 62/117 (52%)
Frame = +1
Query: 313 TVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCF 372
TV +VE K ++ M IW ++ +A + T ++Q TMDR + F++PA S+ F
Sbjct: 52 TVHRVEELKSIIRMGPIWASGILLITAYAQQGTFSLQQAKTMDRHITKSFQIPAGSMSVF 231
Query: 373 IVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQR 429
++T L +YD I RR TG RGI L R+GIG I IA V VE +R
Sbjct: 232 TIITMLTTTALYDRVLIRVARRFTGLDRGISFLHRMGIGFVISTIATFVAGFVEMKR 402
>BP043790
Length = 477
Score = 77.8 bits (190), Expect = 4e-15
Identities = 38/84 (45%), Positives = 56/84 (66%), Gaps = 7/84 (8%)
Frame = -1
Query: 474 FYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKLN-------WIGNNLNDSHLDYY 526
FYDQ+PE MK LGT+ T+ + GS+++TFL++ + + W+ +NLN S LDYY
Sbjct: 477 FYDQAPEGMKSLGTSYFTTSLGLGSFLSTFLLSTVADITKRNGHKGWVLDNLNISRLDYY 298
Query: 527 YAFLFVISALNFGVFLWVSSGYIY 550
YAF+ V+S LNF FL V+ ++Y
Sbjct: 297 YAFMAVLSLLNFLCFLVVAKMFVY 226
>BP064212
Length = 419
Score = 70.5 bits (171), Expect(2) = 5e-15
Identities = 32/39 (82%), Positives = 36/39 (92%)
Frame = -1
Query: 519 NDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTST 557
NDSHL+YY A LFV +ALNFGVFLWVS+GYIY+KENTST
Sbjct: 410 NDSHLEYY*ALLFVKTALNFGVFLWVSNGYIYQKENTST 294
Score = 27.3 bits (59), Expect(2) = 5e-15
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = -3
Query: 562 DIEMSAEKTVKY 573
DIEMSAEKTVKY
Sbjct: 279 DIEMSAEKTVKY 244
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,744,389
Number of Sequences: 28460
Number of extensions: 157696
Number of successful extensions: 1181
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 1143
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1155
length of query: 573
length of database: 4,897,600
effective HSP length: 95
effective length of query: 478
effective length of database: 2,193,900
effective search space: 1048684200
effective search space used: 1048684200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0248.4


