

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0240.1
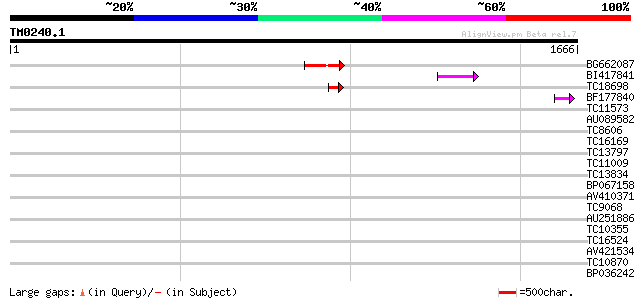
(1666 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG662087 113 3e-25
BI417841 64 3e-10
TC18698 50 4e-06
BF177840 45 9e-05
TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase regi... 39 0.007
AU089582 36 0.043
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 33 0.37
TC16169 similar to GB|BAB08855.1|9758354|AB013396 branched-chain... 32 0.82
TC13797 similar to AAR24648 (AAR24648) At3g53720, partial (10%) 32 1.1
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 31 1.4
TC13834 homologue to UP|Q948R9 (Q948R9) RPS6-like protein (AT3g1... 31 1.8
BP067158 30 3.1
AV410371 29 5.3
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 29 6.9
AU251886 29 6.9
TC10355 homologue to UP|Q8LPD2 (Q8LPD2) Protein kinase Ck2 regul... 29 6.9
TC16524 similar to GB|AAL69537.1|18491137|AY074839 AT5g45420/MFC... 29 6.9
AV421534 28 9.1
TC10870 similar to GB|CAA77438.1|2924285|CHNTXX Ycf2 protein {Ni... 28 9.1
BP036242 28 9.1
>BG662087
Length = 373
Score = 113 bits (282), Expect = 3e-25
Identities = 57/119 (47%), Positives = 80/119 (66%)
Frame = +1
Query: 865 NGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVS 924
+GK R+ +D+ DLN A PKD Y +P + +VD A+ +E LSL+D YSGY+QI + D
Sbjct: 13 SGKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDED 192
Query: 925 KTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSR 983
KTAF A Y + +PFGLKNAGATYQ +M+ +F D + M+VY+D+++VKS R
Sbjct: 193 KTAFMT--ARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKSALR 363
>BI417841
Length = 617
Score = 63.5 bits (153), Expect = 3e-10
Identities = 42/123 (34%), Positives = 64/123 (51%), Gaps = 2/123 (1%)
Frame = +1
Query: 1256 LFFDGSSHKN--GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGA 1313
L FDGSS N G G + + G+ + + + +NN+AEY LI L+ G
Sbjct: 142 LEFDGSSKGNPGSAGAGAVLRAEDGSKVYLREGVGNQ-TNNQAEYRGLILGLKHAHEQGY 318
Query: 1314 RNVVVKGDSELVIKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANEL 1373
+++ VKGDS+LV KQ+ +K + N+A +A L +KF + HV R N EA+
Sbjct: 319 QHINVKGDSQLVCKQVEGSWKARNPNIASLCNEAKELKSKFQSFDINHVPRQYNSEADVQ 498
Query: 1374 AQI 1376
A +
Sbjct: 499 ANL 507
>TC18698
Length = 808
Score = 49.7 bits (117), Expect = 4e-06
Identities = 23/44 (52%), Positives = 32/44 (72%)
Frame = -2
Query: 937 YEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKS 980
Y + VMP GLKN TYQR+M+ IFH I ++VY++D++VKS
Sbjct: 771 YYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKS 640
>BF177840
Length = 410
Score = 45.1 bits (105), Expect = 9e-05
Identities = 23/60 (38%), Positives = 35/60 (58%)
Frame = +2
Query: 1601 SLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLK 1660
S+ +D+ T F+ + G KLL ST + Q +GQ E NKTL +L++++L NLK
Sbjct: 14 SIVSDRDTKFISHFWRTLWGKVGTKLLYSTTCHPQTDGQTEVVNKTLSTLLRSVLERNLK 193
>TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase region
(Fragment), partial (10%)
Length = 572
Score = 38.9 bits (89), Expect = 0.007
Identities = 19/53 (35%), Positives = 29/53 (53%)
Frame = +2
Query: 1605 DQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVE 1657
+ G F ++ F + GI++ S+ + Q NGQ EAANK ++ IK L E
Sbjct: 8 ENGIQFTSKQTQDFCDGMGIQMRFSSVKHPQTNGQTEAANKVILKGIKRRLYE 166
>AU089582
Length = 383
Score = 36.2 bits (82), Expect = 0.043
Identities = 19/63 (30%), Positives = 33/63 (52%)
Frame = +2
Query: 1590 NHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLIS 1649
+ IV G+P S+ +D+G F SF + G +L ST ++ Q +GQ E + L
Sbjct: 38 DEIVSLHGVPVSIISDRGAQFTSHFWRSFQTALGTRLKMSTAFHPQTDGQSERTIQILED 217
Query: 1650 LIK 1652
+++
Sbjct: 218 MLR 226
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 33.1 bits (74), Expect = 0.37
Identities = 19/56 (33%), Positives = 29/56 (50%)
Frame = -2
Query: 346 EKGKDVAEDEDEDMLDDFDDSDGEDLNIICIVSILPAEFDRILEVTENEEDYYDEE 401
E G+D +D+D+D DD +D GED E + +E +NE++ DEE
Sbjct: 416 EDGEDQEDDDDDDEDDDEEDDGGED------------EEEEGVEEEDNEDEEEDEE 285
>TC16169 similar to GB|BAB08855.1|9758354|AB013396 branched-chain amino acid
aminotransferase-like protein {Arabidopsis thaliana;} ,
partial (30%)
Length = 918
Score = 32.0 bits (71), Expect = 0.82
Identities = 23/65 (35%), Positives = 33/65 (50%)
Frame = +2
Query: 1259 DGSSHKNGTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVV 1318
D S N +G G +++SP +P F +KKK +EA + L E L L +N+
Sbjct: 350 DSSVVVNLSGGGTWVISPS*SPVAFWDFLKKKLKQSEAGEDVLDLKSEPLWVL-LKNLRD 526
Query: 1319 KGDSE 1323
KGD E
Sbjct: 527 KGDEE 541
>TC13797 similar to AAR24648 (AAR24648) At3g53720, partial (10%)
Length = 444
Score = 31.6 bits (70), Expect = 1.1
Identities = 15/41 (36%), Positives = 25/41 (60%)
Frame = -2
Query: 345 KEKGKDVAEDEDEDMLDDFDDSDGEDLNIICIVSILPAEFD 385
+E+ + AEDED+D LDD ++ +GE +I + + FD
Sbjct: 326 EEESEGAAEDEDDDGLDD-EEREGEVQRVIALPDSIGGSFD 207
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 31.2 bits (69), Expect = 1.4
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Frame = +2
Query: 348 GKDVAEDEDEDMLDDFDDSDGEDLNIICIV--SILPAEFDRILEVTENEEDYYDEEVPDD 405
G D +D+D+D DD ++ + EDL +V ++ AE + E EED D+ DD
Sbjct: 164 GGDPDDDDDDDDDDDEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDD 343
>TC13834 homologue to UP|Q948R9 (Q948R9) RPS6-like protein
(AT3g17170/K14A17_29), partial (13%)
Length = 517
Score = 30.8 bits (68), Expect = 1.8
Identities = 17/55 (30%), Positives = 31/55 (55%), Gaps = 1/55 (1%)
Frame = +1
Query: 317 RNEPSVASIINQLDHAIKQGTVVPPNPAKEKGKDVAEDEDEDMLDDFDDS-DGED 370
++E + ++ + D AI + PP G V +D+DE++ +D+DD DGE+
Sbjct: 49 QDERVIRHLVVKRDEAITEDCPPPPE-FHTLGASVDDDDDEELDEDYDDDWDGEE 210
>BP067158
Length = 413
Score = 30.0 bits (66), Expect = 3.1
Identities = 14/17 (82%), Positives = 15/17 (87%)
Frame = +3
Query: 30 KGKGRNIVNITTFLVIG 46
+GKGRNIVNIT LVIG
Sbjct: 309 RGKGRNIVNITLPLVIG 359
>AV410371
Length = 347
Score = 29.3 bits (64), Expect = 5.3
Identities = 13/35 (37%), Positives = 20/35 (57%)
Frame = +3
Query: 336 GTVVPPNPAKEKGKDVAEDEDEDMLDDFDDSDGED 370
G++ NP + +D+DEDM++D DD ED
Sbjct: 177 GSLNSNNPNGSNFDNSCQDDDEDMVEDDDDDADED 281
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 28.9 bits (63), Expect = 6.9
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = +2
Query: 341 PNPAKEKGKDVAEDEDEDMLDDFDDSDGE 369
P P + D +D+D+D DD DD DG+
Sbjct: 74 PKPDDDDDDDDDDDDDDDDDDDDDDDDGD 160
>AU251886
Length = 347
Score = 28.9 bits (63), Expect = 6.9
Identities = 9/26 (34%), Positives = 19/26 (72%)
Frame = +1
Query: 426 QRPTEQMKSHLKPLFVWAKVDEKGVN 451
++PT+ M HLKP++V +++K ++
Sbjct: 268 RKPTKDMTKHLKPIYVTTHINDKSMS 345
>TC10355 homologue to UP|Q8LPD2 (Q8LPD2) Protein kinase Ck2 regulatory
subunit 2, partial (25%)
Length = 504
Score = 28.9 bits (63), Expect = 6.9
Identities = 30/128 (23%), Positives = 48/128 (37%), Gaps = 22/128 (17%)
Frame = +2
Query: 269 YSYGSGNYYAPEQMTRTQFRRFLRKRKA--ERERGGKFRSLWDIKP------------ED 314
+ + S + ++ + R + R R+R R+RGG S ++ P +
Sbjct: 53 HQFNSSSSFSESKPYRERERERERERAVIMYRDRGGAVGSKSEVAPPVDRKRINDVLDKQ 232
Query: 315 NPRNEPSVASIINQLDHAIKQGTVV--------PPNPAKEKGKDVAEDEDEDMLDDFDDS 366
R+ PS + IN D A + PP +K E E + D S
Sbjct: 233 LERSSPSTSRPINAKDRASLLSSTAHRDSRNPAPPPISKNSNASDEESETDSEESDVSGS 412
Query: 367 DGEDLNII 374
DGED + I
Sbjct: 413 DGEDTSWI 436
>TC16524 similar to GB|AAL69537.1|18491137|AY074839 AT5g45420/MFC19_9
{Arabidopsis thaliana;}, partial (41%)
Length = 997
Score = 28.9 bits (63), Expect = 6.9
Identities = 35/136 (25%), Positives = 52/136 (37%), Gaps = 8/136 (5%)
Frame = +1
Query: 226 DAGLPKNQWFRPAPPKPKHHQKWSKIDLGQGSSYINNSRVSRSYSYGSGNYYAPEQMTRT 285
D + K Q + KP +W I G + S + +S G
Sbjct: 388 DVEILKKQMAKHPVGKPG---RWEAIAAAFGGRHKAESVIKKSKELGE-----KRSDDSD 543
Query: 286 QFRRFLRKRKAERERGGKFRSLWDIKPEDNPRNEPSVASIINQL-----DHAIKQGTVVP 340
+ +FL+KRKA +RGG ++ E N N +A ++N L D ++ V
Sbjct: 544 SYEQFLKKRKAADKRGGGEEEEGKVEEETNWSNGEDIA-LLNALKAFPKDAPMRWEKVAV 720
Query: 341 PNPAKEKG---KDVAE 353
P K K K VAE
Sbjct: 721 AVPGKSKAACVKRVAE 768
>AV421534
Length = 270
Score = 28.5 bits (62), Expect = 9.1
Identities = 22/81 (27%), Positives = 36/81 (44%), Gaps = 6/81 (7%)
Frame = +3
Query: 284 RTQFRRFLRKRKAERERGGKFRSLWDI---KPEDNPRNEPSVASIINQLDHAIKQGT--- 337
R RR R ERE G + S+ D+ KP+ P P + + + +
Sbjct: 18 RENERRGERDFSKEREFGREASSVGDVSSMKPKTKPFPPPPSSDYHHHRHRSERPSPDRE 197
Query: 338 VVPPNPAKEKGKDVAEDEDED 358
V PP PAK K + +++ + +D
Sbjct: 198 VEPPRPAKRKSEHISDRQRDD 260
>TC10870 similar to GB|CAA77438.1|2924285|CHNTXX Ycf2 protein {Nicotiana
tabacum;}, complete
Length = 6897
Score = 28.5 bits (62), Expect = 9.1
Identities = 13/50 (26%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Frame = +1
Query: 244 HHQKWSKIDLGQGSSYINNSRVSRSYSYGSGNYYAP----EQMTRTQFRR 289
H Q+W + + + + ++ +S SY Y S + + +QMT+T R+
Sbjct: 6685 HRQRWLRTNNSLSNGFFRSNTLSESYQYLSNMFLSNGTLLDQMTKTLLRK 6834
>BP036242
Length = 525
Score = 28.5 bits (62), Expect = 9.1
Identities = 21/74 (28%), Positives = 32/74 (42%)
Frame = +3
Query: 297 ERERGGKFRSLWDIKPEDNPRNEPSVASIINQLDHAIKQGTVVPPNPAKEKGKDVAEDED 356
E E GGK +L + +E D ++ PPN + +D ++DED
Sbjct: 141 ESEEGGKVEALVKQSQTKSSSSE----------DSSVTSTGSYPPNHRYQNCRD-SDDED 287
Query: 357 EDMLDDFDDSDGED 370
+ LDD+ SD D
Sbjct: 288 AEELDDYGASDLSD 329
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,625,023
Number of Sequences: 28460
Number of extensions: 409336
Number of successful extensions: 2647
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 2553
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2634
length of query: 1666
length of database: 4,897,600
effective HSP length: 103
effective length of query: 1563
effective length of database: 1,966,220
effective search space: 3073201860
effective search space used: 3073201860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0240.1


