

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0237.4
(471 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
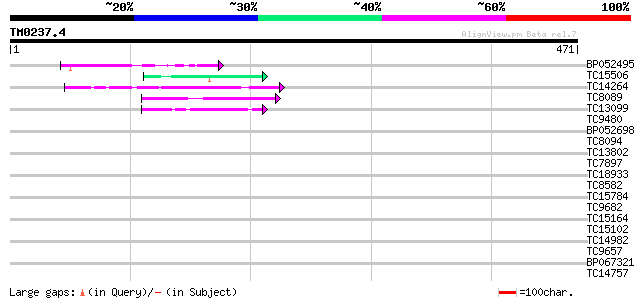
Score E
Sequences producing significant alignments: (bits) Value
BP052495 59 2e-09
TC15506 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich prote... 47 9e-06
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 45 2e-05
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 41 4e-04
TC13099 weakly similar to UP|CBL9_ARATH (Q9FJ13) COBRA-like prot... 41 5e-04
TC9480 weakly similar to UP|Q9AWT3 (Q9AWT3) Nucleoid DNA-binding... 40 0.001
BP052698 40 0.001
TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, c... 39 0.002
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 38 0.004
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 38 0.004
TC18933 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE... 37 0.005
TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 37 0.007
TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precurso... 36 0.012
TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratas... 36 0.012
TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, p... 36 0.015
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 36 0.015
TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%) 36 0.015
TC9657 weakly similar to UP|CAE27414 (CAE27414) 3-hydroxyisobuty... 36 0.015
BP067321 35 0.026
TC14757 similar to UP|O04361 (O04361) Prohibitin, complete 35 0.026
>BP052495
Length = 524
Score = 58.9 bits (141), Expect = 2e-09
Identities = 56/138 (40%), Positives = 66/138 (47%), Gaps = 3/138 (2%)
Frame = -3
Query: 43 VSLADKL---KGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDAT 99
V L D L + RKR P SASQKRAKGAG + S S +Q E + G+
Sbjct: 429 VYLCDHLLLFQSRKRGATPTTSASQKRAKGAGEPSAGNPSPAKSASQREEVQQGGEQGVD 250
Query: 100 IQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGH 159
QA SNP + +P P VE + AKKTS G+R R KSG
Sbjct: 249 EQA-------SNPLLQGTTPIP---------VE-----MAQAKKTS---GKRYSR-KSGS 145
Query: 160 KSPHRSKSSTNSSPSKES 177
S H SKSST+SSP K S
Sbjct: 144 SSSHHSKSSTSSSPLKAS 91
Score = 26.6 bits (57), Expect = 9.4
Identities = 10/15 (66%), Positives = 14/15 (92%)
Frame = -1
Query: 36 DDDEDDNVSLADKLK 50
+DD+DD+V+L DKLK
Sbjct: 524 EDDDDDDVALPDKLK 480
>TC15506 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein, partial
(24%)
Length = 677
Score = 46.6 bits (109), Expect = 9e-06
Identities = 33/113 (29%), Positives = 45/113 (39%), Gaps = 10/113 (8%)
Frame = +2
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHR------- 164
PP R SP P A+S PP+ + + ++ S S RR RS S SP R
Sbjct: 53 PPPRRRSPPPRRARS-------PPRRSPIGRRRSRSPIRRPARSNSRSFSPRRGRPPVRR 211
Query: 165 ---SKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQP 214
S S + SP K S ++ P+R A + PPP +P
Sbjct: 212 GRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPPPPPARKP 370
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 45.4 bits (106), Expect = 2e-05
Identities = 47/184 (25%), Positives = 77/184 (41%), Gaps = 1/184 (0%)
Frame = +2
Query: 46 ADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVH 105
A K K+KP+PAA+ S+ + K A + A+K + A+P K T +A
Sbjct: 470 ASKPAAAKKKPQPAAAKSKPKPK-AKAKPAAAATK--AKAKPAPKSKAAATKTTAKA--- 631
Query: 106 EHSISNPPSRENSPAPNPAKSKAEGVEKPPKA-ASLAKKTSTSEGRRKVRSKSGHKSPHR 164
+ + + P + + PA +KA+ V P KA AS AK + ++ + R P
Sbjct: 632 KPAAAAKPKAKPAAKAKPA-AKAKAVAAPAKAKASPAKPKAKAKSKTAPRMNVNPPVPKA 808
Query: 165 SKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSS 224
++ ++ + T T+P ++ P P PK AT AK G +
Sbjct: 809 KPAAKAKPAARPAKASRTSTRTTPGKKV------PPPPKPAPKKAATPVKKAPAKSGKAK 970
Query: 225 NVSS 228
V S
Sbjct: 971 TVKS 982
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 41.2 bits (95), Expect = 4e-04
Identities = 28/116 (24%), Positives = 47/116 (40%)
Frame = +2
Query: 110 SNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSST 169
S+PPS SP+P G + P ++ T++S+ + H+S S
Sbjct: 263 SSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQ------------NTHQSPPSR 406
Query: 170 NSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSN 225
SSPS S+ +++ A P + P PP++ SP + G S +
Sbjct: 407 TSSPSTSSSTTSAPRSSTRSPTAPPSPPQCTKPPAPPRAPPDSSTSPTSAAGKSDS 574
>TC13099 weakly similar to UP|CBL9_ARATH (Q9FJ13) COBRA-like protein 9
precursor, partial (14%)
Length = 426
Score = 40.8 bits (94), Expect = 5e-04
Identities = 33/105 (31%), Positives = 48/105 (45%)
Frame = +3
Query: 110 SNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSST 169
S+ R PNPA + PP AS ++TS + R R+ S +SP + +ST
Sbjct: 48 SSSSQRRRLRRPNPATVYSSPTPTPP--ASSYRRTSPT--LRSSRTSSSPRSPCSTTAST 215
Query: 170 NSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQP 214
+SSP + S+ T + SP + G P PPP +T P
Sbjct: 216 SSSPGRSSSGFSTMSSWSPPPMRCSPTGL---PSPPPLATVPSSP 341
>TC9480 weakly similar to UP|Q9AWT3 (Q9AWT3) Nucleoid DNA-binding protein
cnd41-like protein, partial (33%)
Length = 716
Score = 39.7 bits (91), Expect = 0.001
Identities = 42/171 (24%), Positives = 60/171 (34%), Gaps = 9/171 (5%)
Frame = -2
Query: 52 RKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISN 111
R R PAA+ SQ +TTS A K +QP T+ H +++
Sbjct: 475 RNRHVSPAAARSQ*-----STATTSLARKQNKTSQPSQPVSGNSSPVTVTPS-HRENVT- 317
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSP--------- 162
PPS + ++S + PP A+ A + T G R+ + P
Sbjct: 316 PPSEPPRSSSTTSRSTCQTTTHPPPPATPAPPS*TRPGVRRTLRSATESPPCRNPQ*FLL 137
Query: 163 HRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQ 213
HR + NS P E P R A G+ P P S T +
Sbjct: 136 HRQRQPRNSPPESE----------EPHRRR*ARSGQGSKTCPDPDSAVTAR 14
>BP052698
Length = 499
Score = 39.7 bits (91), Expect = 0.001
Identities = 30/65 (46%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Frame = +1
Query: 5 QAEVITQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKL---KGRKRKPKPAAS 61
+AE I +P+PQ V ++ V IEDDDDD VSLADKL K +KR P S
Sbjct: 331 KAEPIPRPTPQASPRVTSAT----VEIEDDDDD-----VSLADKLKVXKSQKRGATPNNS 483
Query: 62 ASQKR 66
S KR
Sbjct: 484 XSPKR 498
>TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, chloroplast
precursor (EF-Tu), partial (60%)
Length = 1092
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/103 (27%), Positives = 44/103 (42%)
Frame = +2
Query: 107 HSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSK 166
HS + PPS + P P PA + PP A+S + + + + + + + P
Sbjct: 212 HS*TPPPSSTSPPPPPPAATALPSPSAPPAASSSGRSPT*TSAQSATSTTA--RPP*PPP 385
Query: 167 SSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKST 209
S S PS + + T +T P++ A A PPP ST
Sbjct: 386 SPWRSPPSATAPPRSTTRSTLPLKSAPAA----SPSTPPPSST 502
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 37.7 bits (86), Expect = 0.004
Identities = 43/174 (24%), Positives = 69/174 (38%)
Frame = +2
Query: 55 KPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPS 114
KP S++ + S TS K + P +T++A + ++P S
Sbjct: 80 KPPSQISSNPNKTTSTTSSPTSTTPKPSPSPAPS---------STLRAPSSSPASASPAS 232
Query: 115 RENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPS 174
SP +P K + PP + L T+TSE SP + SS+++SP+
Sbjct: 233 ---SPTKSP-KPSSPSASAPPSSTPLTPSTATSE-----------SSPTATSSSSSASPA 367
Query: 175 KESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSS 228
++ A S E +AL P PP PSP + S+++SS
Sbjct: 368 PPTSF----SALSRALELKALASSPSPPSSPP-------PSPPSAICMSTSLSS 496
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 37.7 bits (86), Expect = 0.004
Identities = 51/188 (27%), Positives = 67/188 (35%), Gaps = 15/188 (7%)
Frame = +3
Query: 56 PKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSR 115
PK + S S K + +G T+G A + G A + A S PPS
Sbjct: 546 PKSSPSPSPKASPPSGSPPTAGEPSPAGTPPSVLPSPAG---APVPAG--GPSAGTPPSA 710
Query: 116 ENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSK 175
SP+P+P V PP A S + G S SP ++T+ SP+
Sbjct: 711 STSPSPSP-------VSAPPAA-------SPAPGAESPSSSPTSNSPAGGPTATSPSPAG 848
Query: 176 ES------ACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPS---------PQAKG 220
+S A T G T P + G D +P P S PS P G
Sbjct: 849 DSPAGGPPAPSPTTGDTPP-----SASGPDVSPSGTPSSLPADTPSSSSNNSTAPPPGNG 1013
Query: 221 GFSSNVSS 228
G S SS
Sbjct: 1014GSSVVPSS 1037
>TC18933 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE11_17
{Arabidopsis thaliana;}, partial (63%)
Length = 609
Score = 37.4 bits (85), Expect = 0.005
Identities = 34/122 (27%), Positives = 50/122 (40%), Gaps = 10/122 (8%)
Frame = +3
Query: 110 SNPPSRENSPAP--NPAKSKAEGVEKPPK-----AASLAKKTSTSEG-RRKVRSKSGHKS 161
S+PPS SP P +KS A+ K P+ +A+ A TST+ VR++ G+ +
Sbjct: 120 SSPPSAATSPKPTNGASKSCAKSAAKSPRFRTKASANTASATSTTRSTNSSVRNRIGNAA 299
Query: 162 PHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPM--PPPKSTATVQPSPQAK 219
S + T + +TSPI A A P P + + P A
Sbjct: 300 SLNSVAPTTLATPPR*LTSTATSSTSPIHPAAAPDTATSAPRKNSPASANSLTNPLSYAS 479
Query: 220 GG 221
GG
Sbjct: 480 GG 485
>TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (65%)
Length = 1341
Score = 37.0 bits (84), Expect = 0.007
Identities = 31/114 (27%), Positives = 44/114 (38%), Gaps = 2/114 (1%)
Frame = +1
Query: 23 SSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQ-KRAKGAGVSTTSGASKL 81
S + ++ D+D DDE D D+ + P PA Q K+ S T + K
Sbjct: 580 SGYEDDLISADEDSDDESD-----DESDDEEETPTPAKKVDQGKKRPNESASKTPVSGKK 744
Query: 82 ASDAQPE-VNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKP 134
A +A PE +GK+ AT + N + SP S G KP
Sbjct: 745 AKNATPEKPDGKKNVHTATPHPKKQDGKTPNSFGKNKSPNSGGKFSNKSGGSKP 906
>TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor (Atperox
P31) (ATP41) , partial (38%)
Length = 475
Score = 36.2 bits (82), Expect = 0.012
Identities = 26/108 (24%), Positives = 38/108 (35%), Gaps = 4/108 (3%)
Frame = +1
Query: 107 HSISNPPSRENSP----APNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSP 162
HS S NSP P+P PP++AS + ST S S
Sbjct: 112 HSTSTNTRAHNSPKSSATPSPTNKSPPPPPAPPRSASSSTTASTPTAATPPSSSPQLPST 291
Query: 163 HRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTA 210
+ + T++SPS + + P P PPP +T+
Sbjct: 292 KPNATPTSTSPSPATPSTSSSELKPPSNSLAPTPSPAPTSSPPPPATS 435
Score = 34.3 bits (77), Expect = 0.045
Identities = 25/113 (22%), Positives = 43/113 (37%), Gaps = 2/113 (1%)
Frame = +1
Query: 106 EHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRS 165
+HS S+PPS ++A PK+++ T+ S +S S S
Sbjct: 70 QHSHSSPPSHTRDSHSTSTNTRAHN---SPKSSATPSPTNKSPPPPPAPPRSASSSTTAS 240
Query: 166 KSSTNSSPSKESACQETQGATSPIREAE--ALPGKDDNPMPPPKSTATVQPSP 216
+ + PS T+ +P + A P + + PP ++ PSP
Sbjct: 241 TPTAATPPSSSPQLPSTKPNATPTSTSPSPATPSTSSSELKPPSNSLAPTPSP 399
>TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (29%)
Length = 731
Score = 36.2 bits (82), Expect = 0.012
Identities = 29/118 (24%), Positives = 46/118 (38%), Gaps = 1/118 (0%)
Frame = +1
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTS-TSEGRRKVRSKSGHKSPHRSKSSTN 170
PP+ N+P P+P S V ++S ++ S + S +PH + + ++
Sbjct: 265 PPNPPNTPDPSPDPSTTSSVNSVSSSSSSESSSAPPSSSSNPLSPASALTTPHLTPTPSS 444
Query: 171 SSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSS 228
PS+ S +T A SP P A P+ QA G +S SS
Sbjct: 445 PLPSQTSTPPQTASAVSP-------------PESAGDGYALSSPAEQASSGATSLTSS 579
>TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, partial
(57%)
Length = 1185
Score = 35.8 bits (81), Expect = 0.015
Identities = 41/129 (31%), Positives = 57/129 (43%), Gaps = 11/129 (8%)
Frame = +2
Query: 110 SNPPSRENSPAPNPAKSKAEGVEKPP--KAASLAKKTSTSEGRRKVRSKSGHKSPHRSKS 167
S PP+ +SP A A KPP A L S S V + S +P S +
Sbjct: 182 STPPT--SSPPATLASLSARSASKPPPPSPARLTPLPSASP----VTTTSTPPTP--SPA 337
Query: 168 STNSSPSKESACQETQGATSPI--------REAEALPGKDDNPMPPPKS-TATVQPSPQA 218
+TN+SPS+ S T TSP + + LPG P+P PK+ +T +P
Sbjct: 338 ATNASPSRRSTTTTTTPPTSPTPMLLTSPPMKLKPLPGY-FLPLPTPKAPISTPLTTPTP 514
Query: 219 KGGFSSNVS 227
K SS++S
Sbjct: 515 KSTPSSSIS 541
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 35.8 bits (81), Expect = 0.015
Identities = 42/190 (22%), Positives = 68/190 (35%), Gaps = 12/190 (6%)
Frame = +1
Query: 56 PKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGD--------ATIQADVHEH 107
P A+AS +TTS + + PE A+ H
Sbjct: 133 PSLEATASPSSEPCTTATTTSTTKAATNSSPPESRSTTPPSSEPTCLLAPASPPTPKSSH 312
Query: 108 SISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKS 167
S + PPSR +S P KSK P A +T+ +P ++
Sbjct: 313 SSTPPPSRSSSTTP---KSKNATSLTAPSCPPPASPAATAP------------APSKTPP 447
Query: 168 STNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTAT----VQPSPQAKGGFS 223
+ ++ SK S+C+ + T+P + A P +P+ P S P A +
Sbjct: 448 NPPTNSSKPSSCKSSLPNTTPSSLSSAPPSPTTSPISTPSSPENQEKPASTPPSAPPRST 627
Query: 224 SNVSSEKLDT 233
S+ SS ++T
Sbjct: 628 SSSSSSPINT 657
>TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%)
Length = 1036
Score = 35.8 bits (81), Expect = 0.015
Identities = 29/113 (25%), Positives = 40/113 (34%)
Frame = +3
Query: 111 NPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTN 170
+PP R NS AP + P AA+ A S S S S S+
Sbjct: 102 SPPQRPNSTAPTALLTALNPSTAAPAAAASAPSAS---------G*SSSSSSSSSSSAVL 254
Query: 171 SSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFS 223
+SPS S T + SP + P PP ++ + +P K S
Sbjct: 255 ASPSTSSTAPTTPPSPSPPSNSPTSTSPPPQPSPPNSTSPSPPQTPTRKTSHS 413
>TC9657 weakly similar to UP|CAE27414 (CAE27414) 3-hydroxyisobutyrate
dehydrogenase , partial (17%)
Length = 765
Score = 35.8 bits (81), Expect = 0.015
Identities = 24/104 (23%), Positives = 49/104 (47%), Gaps = 5/104 (4%)
Frame = -1
Query: 118 SPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKS-GHKSPHRSKSSTN----SS 172
+P P + P+ A + ++ ++ S + +KSP+ S+S ++ +
Sbjct: 384 APEPKQEPEYPSYLHHEPEVTHKAAEQVPNQAKQYPSSDTPSNKSPNPSQSDSHRTLLQN 205
Query: 173 PSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSP 216
PS ES+ + Q ++P R ++ PGK +P T ++ P+P
Sbjct: 204 PSSESSSRTNQATSNPPRRWKSRPGKAPHPALLSARTPSLPPTP 73
>BP067321
Length = 513
Score = 35.0 bits (79), Expect = 0.026
Identities = 30/121 (24%), Positives = 42/121 (33%), Gaps = 4/121 (3%)
Frame = +1
Query: 112 PPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNS 171
PP E + P + PP L +++ + + K+ H SST +
Sbjct: 28 PPFLEGGTSSPPYLQRHHPTSPPPPPPPLRRQSQSPNTHLQKNPKTRHHYRRTRASSTTT 207
Query: 172 SPSKES----ACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVS 227
PS S A Q AT P P S+AT +P Q+ F S S
Sbjct: 208 IPSTSSPWNAAASSRQTATGP*SSGSPACRTSSPAPPSSSSSATRRPRRQSPWRFGSRRS 387
Query: 228 S 228
S
Sbjct: 388 S 390
>TC14757 similar to UP|O04361 (O04361) Prohibitin, complete
Length = 1141
Score = 35.0 bits (79), Expect = 0.026
Identities = 29/123 (23%), Positives = 47/123 (37%), Gaps = 9/123 (7%)
Frame = +1
Query: 110 SNPPSRENS-PAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSS 168
S PP + + A P ++ PP A++L ST + + + S + SS
Sbjct: 10 SGPPRQISKWVATKPQSPSSQTSPAPPSASALPPPPSTPPSTPSMEASAPSSSTDSAASS 189
Query: 169 TNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPK--------STATVQPSPQAKG 220
T +PS + + G+ +P AL P P P+ S + P+P A
Sbjct: 190 T--TPSAKEPISSSHGSRNPTSSTSALAPTPSPPSPAPRISRW*TSPSVFSPAPTPNASP 363
Query: 221 GFS 223
S
Sbjct: 364 SLS 372
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.307 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,147,497
Number of Sequences: 28460
Number of extensions: 87267
Number of successful extensions: 1790
Number of sequences better than 10.0: 245
Number of HSP's better than 10.0 without gapping: 1251
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1529
length of query: 471
length of database: 4,897,600
effective HSP length: 94
effective length of query: 377
effective length of database: 2,222,360
effective search space: 837829720
effective search space used: 837829720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0237.4


