

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.5
(406 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
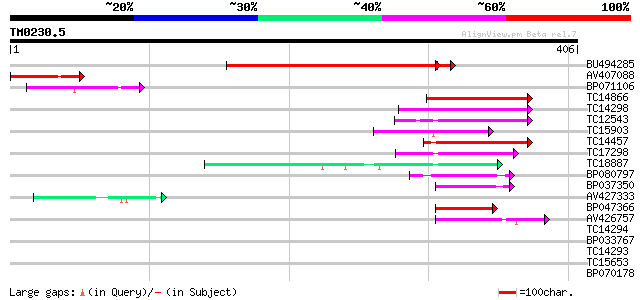
Score E
Sequences producing significant alignments: (bits) Value
BU494285 315 8e-88
AV407088 84 4e-17
BP071106 78 2e-15
TC14866 similar to UP|Q8L5W2 (Q8L5W2) bZIP transcription factor ... 65 3e-11
TC14298 homologue to UP|Q8L5W2 (Q8L5W2) bZIP transcription facto... 61 3e-10
TC12543 similar to UP|Q9AT29 (Q9AT29) bZIP transcription factor,... 61 4e-10
TC15903 similar to UP|Q40724 (Q40724) Transcriptional activator ... 59 1e-09
TC14457 similar to UP|Q8GTZ1 (Q8GTZ1) bZIP transcription factor,... 57 4e-09
TC17298 similar to UP|Q9AT29 (Q9AT29) bZIP transcription factor,... 54 4e-08
TC18887 similar to UP|Q94KA6 (Q94KA6) bZIP transcription factor ... 52 2e-07
BP080797 47 6e-06
BP037350 44 4e-05
AV427333 44 4e-05
BP047366 44 5e-05
AV426757 41 3e-04
TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 40 0.001
BP033767 39 0.001
TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 39 0.002
TC15653 similar to UP|Q941L5 (Q941L5) bZIP transcription factor ... 38 0.003
BP070178 37 0.006
>BU494285
Length = 504
Score = 315 bits (808), Expect(2) = 8e-88
Identities = 151/153 (98%), Positives = 153/153 (99%)
Frame = +1
Query: 156 KNNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGT 215
+NNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGT
Sbjct: 13 RNNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGT 192
Query: 216 AVHTTQNGGLNTPHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNV 275
AVHTTQNGGLNTPHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNV
Sbjct: 193 AVHTTQNGGLNTPHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNV 372
Query: 276 PSTAVAGGMATGGSRDGVQSQHWLQDERELKRQ 308
PSTAVAGGMATGGSRDGVQSQHWLQDEREL+RQ
Sbjct: 373 PSTAVAGGMATGGSRDGVQSQHWLQDERELQRQ 471
Score = 25.4 bits (54), Expect(2) = 8e-88
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +3
Query: 307 RQRRKQSNRESAR 319
+ RRKQSNRESAR
Sbjct: 465 KTRRKQSNRESAR 503
>AV407088
Length = 386
Score = 84.0 bits (206), Expect = 4e-17
Identities = 39/53 (73%), Positives = 43/53 (80%)
Frame = +1
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGF 53
MGSS+ DK PKEKESKTPP T QEQ PTT G VN PDW++FQAYS +PPHGF
Sbjct: 229 MGSSDADKPPKEKESKTPPSTSQEQSPTTGAGPVN-PDWANFQAYSHIPPHGF 384
>BP071106
Length = 402
Score = 78.2 bits (191), Expect = 2e-15
Identities = 42/91 (46%), Positives = 54/91 (59%), Gaps = 7/91 (7%)
Frame = +3
Query: 13 KESKTPPLTPQEQPPTTSTGTVNPPDWSS-FQAY-----SPMPPHGFLASNPQAHPYMWG 66
+ES P P T + T + PDWSS QAY +P P + ++P HPY+WG
Sbjct: 123 EESMDKPSKPSSTSSTQISPTPSYPDWSSSMQAYYAPGVTPPPFYASTVASPAIHPYIWG 302
Query: 67 VQH-IMPPYGTPPHPYVAMYPHGGIYAHPSI 96
QH ++PPYGTP PY AMYP G +YAHPS+
Sbjct: 303 GQHPLIPPYGTPV-PYPAMYPPGAVYAHPSM 392
>TC14866 similar to UP|Q8L5W2 (Q8L5W2) bZIP transcription factor ATB2,
partial (60%)
Length = 948
Score = 64.7 bits (156), Expect = 3e-11
Identities = 30/76 (39%), Positives = 50/76 (65%)
Frame = +3
Query: 299 LQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYE 358
LQ + ++ +RKQSNRESARRSR+RKQ+ ++L + L +EN + + ++ Y+
Sbjct: 36 LQMVMDQRKNKRKQSNRESARRSRMRKQSHLEDLTSQVTQLTKENGEILTNINITSQQYQ 215
Query: 359 QLLTENAALKERLGEL 374
+ TEN+ L+ ++GEL
Sbjct: 216 NVETENSILRAQMGEL 263
>TC14298 homologue to UP|Q8L5W2 (Q8L5W2) bZIP transcription factor ATB2,
partial (62%)
Length = 1585
Score = 61.2 bits (147), Expect = 3e-10
Identities = 35/97 (36%), Positives = 57/97 (58%), Gaps = 1/97 (1%)
Frame = +3
Query: 279 AVAGGMATGGSR-DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAE 337
A + G ++G S S+ LQ + ++++R SNRESARRSR+RKQ D+LA +
Sbjct: 615 ASSSGTSSGSSLLQNSGSEEDLQAVMDQRKRKRMISNRESARRSRMRKQKHLDDLASQVT 794
Query: 338 ALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
L++EN + + V+ Y + EN+ L+ ++GEL
Sbjct: 795 QLRKENQQIITSVNITTQQYLSVEAENSVLRAQMGEL 905
>TC12543 similar to UP|Q9AT29 (Q9AT29) bZIP transcription factor, partial
(52%)
Length = 510
Score = 60.8 bits (146), Expect = 4e-10
Identities = 39/99 (39%), Positives = 54/99 (54%)
Frame = +3
Query: 276 PSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQR 335
P ++ +T D + Q L +ER + RR SNRESARRSR+RKQ DEL +
Sbjct: 162 PQSSCISSNSTSDEAD--EQQQGLINER---KHRRMISNRESARRSRMRKQRHLDELWSQ 326
Query: 336 AEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
L+ EN L ++S ++Q++ ENA LKE EL
Sbjct: 327 VVWLRNENHQLLDKLSHASESHDQVVQENAQLKEEAIEL 443
>TC15903 similar to UP|Q40724 (Q40724) Transcriptional activator protein
(RITA-1 protein), partial (21%)
Length = 603
Score = 58.9 bits (141), Expect = 1e-09
Identities = 37/88 (42%), Positives = 49/88 (55%), Gaps = 2/88 (2%)
Frame = +1
Query: 261 GTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDE--RELKRQRRKQSNRESA 318
G+P S+N P + G + G + D ++ Q +++KR RRK SNRESA
Sbjct: 325 GSPVSANKPNMGGENHLKGTSSGSSDRSDEDDDEAGPCEQSNNPQDVKRLRRKVSNRESA 504
Query: 319 RRSRLRKQAECDELAQRAEALKEENASL 346
RRSR RKQA EL + E LK ENA+L
Sbjct: 505 RRSRRRKQAHLAELETQVEKLKLENATL 588
>TC14457 similar to UP|Q8GTZ1 (Q8GTZ1) bZIP transcription factor, partial
(50%)
Length = 645
Score = 57.4 bits (137), Expect = 4e-09
Identities = 30/78 (38%), Positives = 48/78 (61%)
Frame = +1
Query: 297 HWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSD 356
H + D+ K+++R QSNRESARRSR+RKQ + ++ + E LK+EN + + +
Sbjct: 1 HHVMDQ---KKRKRMQSNRESARRSRMRKQEHLEGMSAQVEQLKKENNQISTNIGVTTQM 171
Query: 357 YEQLLTENAALKERLGEL 374
Y + ENA L+ ++ EL
Sbjct: 172 YLNVEAENAILRVQMAEL 225
>TC17298 similar to UP|Q9AT29 (Q9AT29) bZIP transcription factor, partial
(39%)
Length = 484
Score = 54.3 bits (129), Expect = 4e-08
Identities = 30/88 (34%), Positives = 49/88 (55%)
Frame = +1
Query: 277 STAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRA 336
S+ ++ + + D Q Q + +ER + RR SNRESARRSR+RKQ DEL
Sbjct: 229 SSCISSNSTSDEAEDQQQQQSLIINER---KHRRMISNRESARRSRMRKQKHLDELWSMV 399
Query: 337 EALKEENASLRSEVSRIRSDYEQLLTEN 364
L+ EN L +++ + +++++ EN
Sbjct: 400 VCLRNENHQLLEKLNHLSESHDRVVQEN 483
>TC18887 similar to UP|Q94KA6 (Q94KA6) bZIP transcription factor 6, partial
(64%)
Length = 944
Score = 52.0 bits (123), Expect = 2e-07
Identities = 63/226 (27%), Positives = 91/226 (39%), Gaps = 12/226 (5%)
Frame = +3
Query: 140 IKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKS 199
+ R G +G N I +++ + S N + +G + +++ QN + +
Sbjct: 276 MNRIAGLMGGNNRIPGASDDPIVSLQNSTNLPLNANGFRSPNQQQQMQNSHQQNHQQQQH 455
Query: 200 GGRRDSFEDEPSQNGTAVHTTQN--GGLNTPHQTMSMVPISA-----GGAPGAVPGPATN 252
++ F +P+ N Q GG+ M A GG G PGP
Sbjct: 456 HQQQQIFPKQPALNYATQMPNQGIRGGIVGLSADQGMNGNLAQGGGIGGLVGLAPGPVQL 635
Query: 253 LNIGMDYWGTP----TSSNIPAL-HGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKR 307
+ G+P TSS+I +G+ S + + GG R ER R
Sbjct: 636 AS------GSPANQMTSSDIMGKSNGDTSSVSPVPYVFNGGLRGRKTGAVEKVIER---R 788
Query: 308 QRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRI 353
QRR NRESA RSR RKQA EL Q LKEEN L+ + I
Sbjct: 789 QRRMIKNRESAARSRARKQAYTMELEQEVAKLKEENEELQKKQEEI 926
>BP080797
Length = 498
Score = 47.0 bits (110), Expect = 6e-06
Identities = 32/76 (42%), Positives = 43/76 (56%), Gaps = 1/76 (1%)
Frame = -2
Query: 287 GGSRDGVQSQHWLQDERELKRQ-RRKQSNRESARRSRLRKQAECDELAQRAEALKEENAS 345
G RDG + E+ L+R+ RRK NRESA RSR RKQA +EL + L+++N
Sbjct: 491 GRKRDGPDAY-----EKALERRLRRKIKNRESAARSRARKQAYHNELVSKVTRLEQDNIK 327
Query: 346 LRSEVSRIRSDYEQLL 361
L+ E D+EQ L
Sbjct: 326 LKKE-----KDFEQSL 294
>BP037350
Length = 495
Score = 44.3 bits (103), Expect = 4e-05
Identities = 26/56 (46%), Positives = 33/56 (58%)
Frame = -2
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLL 361
+RQ+R NRESA RSR RK A +EL + L+EEN LR R + EQ+L
Sbjct: 374 RRQKRMIKNRESAARSRARKHAYTNEL*NKVSRLEEENERLRK-----RKELEQML 222
>AV427333
Length = 387
Score = 44.3 bits (103), Expect = 4e-05
Identities = 34/107 (31%), Positives = 40/107 (36%), Gaps = 12/107 (11%)
Frame = +3
Query: 18 PPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQHIMPPYGTP 77
PP + PPT + NPP S SP PP A P+ HP PP +P
Sbjct: 84 PPPSHPYNPPTPPSHPFNPPPTPSHYFKSPPPPSHSFAPPPRGHP--------SPPPSSP 239
Query: 78 P----HPYV--------AMYPHGGIYAHPSIPPGSYPFSPFAMPSPN 112
P HP+ PH I P + P P P PSPN
Sbjct: 240 PPPSSHPFTPPPPHVRPPPSPHHPITPPPHVRPPPPPLPP--SPSPN 374
Score = 35.4 bits (80), Expect = 0.017
Identities = 28/99 (28%), Positives = 39/99 (39%), Gaps = 8/99 (8%)
Frame = +3
Query: 42 FQAYSPMPPHGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYA-----HPSI 96
F + P P H + P +HP+ P TP H + + P +A HPS
Sbjct: 69 FPHFPPPPSHPYNPPTPPSHPF--------NPPPTPSHYFKSPPPPSHSFAPPPRGHPSP 224
Query: 97 PPGSYP---FSPFAMPSPNGIVEASGNTPGSMEADGKPP 132
PP S P PF P P+ S + P + +PP
Sbjct: 225 PPSSPPPPSSHPFTPPPPHVRPPPSPHHPITPPPHVRPP 341
>BP047366
Length = 535
Score = 43.9 bits (102), Expect = 5e-05
Identities = 23/44 (52%), Positives = 28/44 (63%)
Frame = -3
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSE 349
+RQ+R NRESA RSR RKQA EL + L+EEN LR +
Sbjct: 485 RRQKRMIKNRESAARSRARKQAYTQELEIKVSRLEEENERLRRQ 354
>AV426757
Length = 415
Score = 41.2 bits (95), Expect = 3e-04
Identities = 26/83 (31%), Positives = 47/83 (56%), Gaps = 2/83 (2%)
Frame = +1
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLL--TE 363
KR +R +NR+SA+RSR+RK EL + L+ E ++L V+ + D+++L+ +
Sbjct: 46 KRVKRILANRQSAQRSRVRKLQYISELERSVTTLQTEVSALSPRVAFL--DHQRLILNVD 219
Query: 364 NAALKERLGELPGNDDLRSGKSD 386
N+ LK+R+ L + + D
Sbjct: 220 NSTLKQRIAALAQDKIFKDAHQD 288
>TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (65%)
Length = 1192
Score = 39.7 bits (91), Expect = 0.001
Identities = 36/123 (29%), Positives = 44/123 (35%), Gaps = 8/123 (6%)
Frame = +1
Query: 10 PKEKESKTPPLTPQ-EQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQ 68
P + PP P PP+ S G PP Q Y PP + P + Y
Sbjct: 112 PYQPYGAAPPSQPYGAAPPSQSYGAPPPP-----QPYGAAPPSQPYGAPPPSQSYGGPPP 276
Query: 69 HIMP----PYGTPPHPYVAMY---PHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNT 121
P PYG P PY A Y P ++H YP P A SP + S
Sbjct: 277 PSQPYSASPYGQPSAPYAAPYQKPPKDESHSHGGGGGSGYPPPPSAYGSPFASLVPSVFP 456
Query: 122 PGS 124
PG+
Sbjct: 457 PGT 465
>BP033767
Length = 541
Score = 39.3 bits (90), Expect = 0.001
Identities = 29/112 (25%), Positives = 46/112 (40%), Gaps = 3/112 (2%)
Frame = +1
Query: 24 EQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVA 83
+ P +T + NP + F +P H + P++ PP PP P +
Sbjct: 118 QNPSSTGGSSTNPSHKAPFHPLNPQTHH-----HHHHLPHLQTQNTHFPPPPQPPPPKMP 282
Query: 84 MY---PHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEADGKPP 132
+ PH ++ H +PP S P P + PS + + + N P S A PP
Sbjct: 283 PHLSSPHTHLHHHLLLPPPSPPSPPTSPPS-SSLKPPNPNPPTSSSASPLPP 435
>TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (18%)
Length = 540
Score = 38.9 bits (89), Expect = 0.002
Identities = 35/115 (30%), Positives = 42/115 (36%), Gaps = 8/115 (6%)
Frame = +2
Query: 18 PPLTPQ-EQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQHIMP---- 72
PP P PP+ S G PP Q Y PP + P + Y P
Sbjct: 149 PPYQPYGAAPPSQSYGAPPPP-----QPYGAAPPSQPYGAPPPSQSYGGPPPPSQPYSAS 313
Query: 73 PYGTPPHPYVAMY---PHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGS 124
PYG P PY A Y P ++H YP P A SP + S PG+
Sbjct: 314 PYGQPSAPYAAPYQKPPKDESHSHGGGGGSGYPPPPSAYGSPFASLVPSVFPPGT 478
>TC15653 similar to UP|Q941L5 (Q941L5) bZIP transcription factor BZI-4,
partial (25%)
Length = 511
Score = 38.1 bits (87), Expect = 0.003
Identities = 25/60 (41%), Positives = 34/60 (56%), Gaps = 2/60 (3%)
Frame = +2
Query: 283 GMAT--GGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALK 340
GMA+ GS G + D+R+ KR SNRESARRSR+RKQ + ++L L+
Sbjct: 338 GMASVQRGSSSGSEGGDLAIDDRKRKRM---VSNRESARRSRMRKQKQLEDLTAEMSRLQ 508
>BP070178
Length = 385
Score = 37.0 bits (84), Expect = 0.006
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = -2
Query: 391 DDAQQSGQTEAVQGGH 406
DDAQQSGQTEAVQGGH
Sbjct: 384 DDAQQSGQTEAVQGGH 337
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.306 0.126 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,148,437
Number of Sequences: 28460
Number of extensions: 139225
Number of successful extensions: 1134
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 985
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1079
length of query: 406
length of database: 4,897,600
effective HSP length: 92
effective length of query: 314
effective length of database: 2,279,280
effective search space: 715693920
effective search space used: 715693920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0230.5


