

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0228a.4
(1904 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
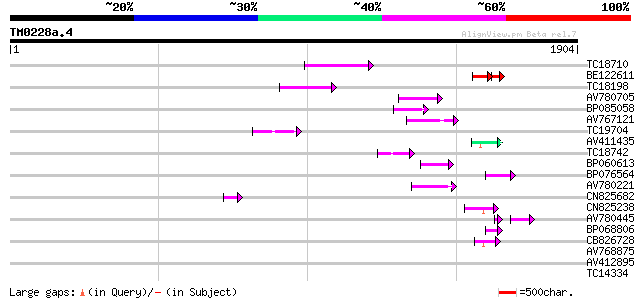
Score E
Sequences producing significant alignments: (bits) Value
TC18710 151 8e-37
BE122611 99 2e-31
TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, p... 119 3e-27
AV780705 83 4e-16
BP085058 83 5e-16
AV767121 77 2e-14
TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA po... 77 3e-14
AV411435 58 2e-08
TC18742 similar to UP|Q9FZG5 (Q9FZG5) T2E6.4, partial (3%) 58 2e-08
BP060613 57 2e-08
BP076564 55 8e-08
AV780221 53 4e-07
CN825682 50 4e-06
CN825238 50 4e-06
AV780445 41 6e-06
BP068806 45 1e-04
CB826728 43 5e-04
AV768875 42 0.001
AV412895 41 0.002
TC14334 weakly similar to UP|AMPL_YEAST (P14904) Vacuolar aminop... 40 0.004
>TC18710
Length = 843
Score = 151 bits (382), Expect = 8e-37
Identities = 84/232 (36%), Positives = 128/232 (54%), Gaps = 1/232 (0%)
Frame = +2
Query: 990 EEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLV 1049
+++R +++ + G DG + FY++ ++ + + + + + E+ TL+
Sbjct: 140 KKLRLLFFLLETSQPRGMDGMNGLFYQKNGDVVKDSVT*AILAFLERAEIPNEINETLVT 319
Query: 1050 LIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNI 1109
LIPKV +P + RPIS C YK+I+K+ V R + + L+SP+Q FI GR +DN+
Sbjct: 320 LIPKVPHPESINQFRPISGCTFLYKVISKIFVARLKDAMPDLISPMQSGFIQGRQIQDNL 499
Query: 1110 ILAQEVMHTIH-TRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGV 1168
++ QE H I+ G +K+D+ KAYDRV W FL ++L FGF V +IM V
Sbjct: 500 LIVQEAFHAINRPGALGRNHSIIKLDMNKAYDRVEWKFLESSLLAFGFSTNWVKMIMILV 679
Query: 1169 RTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEG 1220
+ + NG P QRGLRQGDP SPYLF+ ME L++ IQN G
Sbjct: 680 SGVSYNYKINGVVGPKLLPQRGLRQGDPFSPYLFLFTMEVLSLLIQNSFNMG 835
>BE122611
Length = 357
Score = 99.4 bits (246), Expect(2) = 2e-31
Identities = 45/66 (68%), Positives = 52/66 (78%)
Frame = -3
Query: 1555 GEDRILWREASDDIYSASSAYQFLNEDGLPETGFWKLIWKASAPEKVRFFLWLVGRDALP 1614
G DRILW E S+ YSA S Y L EDG E+ FW L+WKA+APEKVRFFLWLVGR+ALP
Sbjct: 337 GTDRILWTEESNGCYSAKSTYNLLIEDGGNESRFWSLVWKANAPEKVRFFLWLVGRNALP 158
Query: 1615 TNAKRH 1620
+NA+RH
Sbjct: 157 SNARRH 140
Score = 55.8 bits (133), Expect(2) = 2e-31
Identities = 24/42 (57%), Positives = 30/42 (71%)
Frame = -2
Query: 1618 KRHRHHLATTASCIRCGAVVEDAEHVLRRCPGSVWVRGQLGR 1659
K+ +HLA++A+CIRCGA EDA HV R CPGS W+ Q R
Sbjct: 149 KKAPYHLASSAACIRCGAPSEDANHVFRWCPGSAWMWTQFSR 24
>TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, partial (14%)
Length = 592
Score = 119 bits (299), Expect = 3e-27
Identities = 63/189 (33%), Positives = 102/189 (53%)
Frame = -3
Query: 907 GDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQVRRNR 966
GD NT+FFHA T+ RR N+I + G W + E A ++++++S
Sbjct: 569 GDHNTRFFHASTIQRRDFNRILKIKDAHGIWVEGQKKVNEAAVNYFSDIYSSSPLQYLRE 390
Query: 967 IMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDL 1026
+ + KL A VS +E++ A+ SM KAPG DG + FY+Q I+ D+
Sbjct: 389 CLAPIPKLVTDRLNHKLCAQVSTDEIKEAVFSMGDLKAPGMDGINGLFYQQNREIVKNDV 210
Query: 1027 HHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRP 1086
++ + F +G EL TL+ LIPK+ + ++ RPIS C+ YK+I+K++V R +P
Sbjct: 209 NNAILDFFDHGELPVELNETLVTLIPKIPHAEAIQQFRPISCCSFIYKVISKIIVARLKP 30
Query: 1087 LLHKLVSPL 1095
+ L+SP+
Sbjct: 29 DMVNLISPM 3
>AV780705
Length = 524
Score = 83.2 bits (204), Expect = 4e-16
Identities = 49/149 (32%), Positives = 72/149 (47%), Gaps = 1/149 (0%)
Frame = -2
Query: 1306 GKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQS 1365
G YLGLP + GR K I EKV +L WK LLN+AG+ L K+I+ ++P +AM
Sbjct: 490 GHYLGLPAIWGRNKSHSLVWIEEKVKEKLEGWKETLLNQAGKEVLIKAIIQAIPSYAMTI 311
Query: 1366 LWLPDSVCNHVDKMVRNCIW-GKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALL 1424
+ P + CN + +V + W +G A + SK KG +G R R N A L
Sbjct: 310 VHFPKTFCNSLYALVADFWWKSQGEGAVEYTGAVGTK*PLSKEKGGVGFRDLRTQNLAFL 131
Query: 1425 GKLVHSFLNEKGKLWVQALSQKYIKSGSI 1453
+ L LWV+ + Y + ++
Sbjct: 130 ARQAWRVLTNPEALWVRVMKSLYFPTPAL 44
>BP085058
Length = 452
Score = 82.8 bits (203), Expect = 5e-16
Identities = 44/119 (36%), Positives = 70/119 (57%)
Frame = +2
Query: 1289 RQRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRV 1348
R R +S + I N+GKY+ P+ QG V ++ F +++V +RLA K LLNK R+
Sbjct: 83 RDRMVS-ITSIPFTDNIGKYMSFPIHQGHV*KEDFVFTLDRVISRLAC*KAYLLNKPARL 259
Query: 1349 CLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKA 1407
LAK +LS++ ++ MQ WLP S+C+ + +VR+ +W N LVAW+ + K+
Sbjct: 260 TLAKPVLSAISMYVMQLNWLPQSICDQICTVVRHFVW---ENVNGIPLVAWETIQSRKS 427
>AV767121
Length = 525
Score = 77.4 bits (189), Expect = 2e-14
Identities = 52/178 (29%), Positives = 82/178 (45%), Gaps = 6/178 (3%)
Frame = +2
Query: 1334 LASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARS 1393
L+ WK ++ GR+ L +S+L++LP+ + LP V +++RN +WG N
Sbjct: 2 LSKWKQKTVSFGGRIFLIQSVLTALPLFFLSFFKLPIGVGKSCVRLMRNFLWGGSENENK 181
Query: 1394 WHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGKLWVQALSQKYIKSGSI 1453
V W DV + K G LG++ N+ALLGK +L E LW + + +
Sbjct: 182 IA*VKWTDVCKPKELGGLGIKDLFTFNKALLGK*RWRYLTEPDSLWRRVIEAQ------- 340
Query: 1454 LTEELRPGGSYVWRGILK-----ARDSVSDGFGPRLGAG-SSSLWYSNWLGTGKIASR 1505
+ GS W IL A S G +G G + W +WLG+G +++R
Sbjct: 341 --PDHYSCGSSWWNDILSLCPEDADGWFSSGLKKLVGEGDQTKFWSEDWLGSGLLSAR 508
>TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA
polymerase-like protein, partial (3%)
Length = 530
Score = 77.0 bits (188), Expect = 3e-14
Identities = 51/172 (29%), Positives = 79/172 (45%), Gaps = 8/172 (4%)
Frame = -3
Query: 815 NTSWRAQD--GNLQDKLQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQE----HDWR 868
NTS A NL K + A TE++ F + ++ +L+ +Q DW+
Sbjct: 495 NTSETASSHCSNLSGKTTNTKLALTEWHHQNFSRADKEIAELKNQLEHLQNYSNSGEDWQ 316
Query: 869 DSESLARLERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIH 928
+ + +DL + QEE+ W Q+SR W++ GD+NTKFFHA TV RR+RN+I
Sbjct: 315 EIKQRKMRLKDLWR-------QEELFWSQRSRIKWLKSGDKNTKFFHASTVQRRERNRIE 157
Query: 929 GLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQVRRNRIME--NQTPTIPME 978
+ G W + + FY +++ R I E N P + E
Sbjct: 156 RIKDMQGTWVRSQLAINRDVPDFYPDIYK---STNRQNITECLNAVPRVIYE 10
>AV411435
Length = 426
Score = 57.8 bits (138), Expect = 2e-08
Identities = 34/115 (29%), Positives = 44/115 (37%), Gaps = 11/115 (9%)
Frame = +3
Query: 1551 SIQTGEDRILWREASDDIYSASSAYQFL-----------NEDGLPETGFWKLIWKASAPE 1599
SI EDR+ W YS S Y+ + + P + W IW A P+
Sbjct: 21 SITGSEDRLFWPFIPSGEYSVKSGYRAVKATQFNLSNLASSSSHPSSFLWHTIWGAQVPK 200
Query: 1600 KVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPGSVWVR 1654
KVR FLW +A+P R ++ C C ED H L C W R
Sbjct: 201 KVRSFLWRAANNAIPVKRNLKRRNMGRDDFCPICNKGPEDINHALLACE---WTR 356
>TC18742 similar to UP|Q9FZG5 (Q9FZG5) T2E6.4, partial (3%)
Length = 912
Score = 57.8 bits (138), Expect = 2e-08
Identities = 34/125 (27%), Positives = 65/125 (51%)
Frame = -3
Query: 1234 ISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQREL 1293
ISHL FAD++++F + + +I + L F SG+ N KS + +V + + ++
Sbjct: 373 ISHLCFADNLMVFSNGYLESIAIINNVLHIFQHLSGLTPNPAKSEVFI--LVKETTKNQI 200
Query: 1294 SGLVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKS 1353
+ ++G + LG+P L ++K ++++ ++L W L+ AGR+ L S
Sbjct: 199 TNMLGYKEGKLPVR*LGVPQLTTKLKASDCKVLVDRKPSKLQHWTGRSLSYAGRLQLINS 20
Query: 1354 ILSSL 1358
IL S+
Sbjct: 19 ILFSM 5
>BP060613
Length = 378
Score = 57.4 bits (137), Expect = 2e-08
Identities = 34/112 (30%), Positives = 49/112 (43%), Gaps = 1/112 (0%)
Frame = +1
Query: 1379 MVRNC-IWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGK 1437
M+ C +W R H W +T+ K +G LG + N+ALL K L+
Sbjct: 25 MLSGCQVWWSSKGTRGVHWRKWDLLTELKDEGGLGFKDFEIQNQALLAKQAWRILHNPDA 204
Query: 1438 LWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS 1489
LWVQ L Y L R G S+VW +L R+ + +LG+G +
Sbjct: 205 LWVQILKALYFPHHDFLQTTKRTGASWVWSSLLHGRELLLKQGQWQLGSGET 360
>BP076564
Length = 371
Score = 55.5 bits (132), Expect = 8e-08
Identities = 34/103 (33%), Positives = 49/103 (47%), Gaps = 2/103 (1%)
Frame = +2
Query: 1599 EKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPGS--VWVRGQ 1656
EK +F + L ALP NA+RH +A T C RC A V+D H L+ CP S VW R
Sbjct: 68 EKCKFMVGLGLHQALPINARRHACSMAATGGCGRCSAAVKDILHCLQDCPHSREVWNRLG 247
Query: 1657 LGRLLPWISDSATFRHWLFGHLQNRDNTLFCAMAWNLWKWRNS 1699
++S WL ++ + + W +W+ RN+
Sbjct: 248 CSSFGGFLSSDGW--SWLLSIMKQPNVEKLIYILWWIWR*RNN 370
>AV780221
Length = 461
Score = 53.1 bits (126), Expect = 4e-07
Identities = 49/167 (29%), Positives = 75/167 (44%), Gaps = 16/167 (9%)
Frame = +3
Query: 1348 VCLAKSILSSLPVHAMQSLWLPD-----SVCNHVDKMVRNCIWGKGGNARSWHLVAWKDV 1402
+CL +S+L +LP+ + LP +V N D + +GG +W V W V
Sbjct: 3 LCLIRSVLIALPLFHLPFFKLPTGGFYTNVNN**DPFFGREV--EGGKKIAW--VKWSTV 170
Query: 1403 TQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGKLWVQALSQKY----IKSGSILTEEL 1458
+ K +G LG+++ N+ALLGK L E+ LW + L KY +S S +L
Sbjct: 171 CRPKDEGGLGVQNLGLFNKALLGKWRWRMLKERDGLWYKVLLIKYKNAIPQSASSWWNDL 350
Query: 1459 RP------GGSYVWRGILKARDSVSDGFGPRLGAGSS-SLWYSNWLG 1498
GG ++ RG+ + R+G G+ W NWLG
Sbjct: 351 HSTCFEDGGGGWMQRGLCR-----------RIGEGTEVKFWNENWLG 458
>CN825682
Length = 626
Score = 49.7 bits (117), Expect = 4e-06
Identities = 24/64 (37%), Positives = 33/64 (51%)
Frame = -3
Query: 718 NMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYS 777
+ L+ N +DL G +FTW + +RLDR L + WRL +P A L + S
Sbjct: 207 DFLDGANLMDLELKGCRFTWLSNLRNGLVTKERLDRVLVNWTWRLDYPNATAMALPIISS 28
Query: 778 DHSP 781
DHSP
Sbjct: 27 DHSP 16
>CN825238
Length = 692
Score = 49.7 bits (117), Expect = 4e-06
Identities = 29/121 (23%), Positives = 51/121 (41%), Gaps = 7/121 (5%)
Frame = +3
Query: 1527 WNFDRLYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLNEDGLPET 1586
WN + L P+E I S+ + +T + LWR + + +S SAY + +T
Sbjct: 321 WNVQLIDQLFPKETAAVIKSIPL-GWRTTRNSCLWRWSRNGAFSVRSAYYNIQVAKEMQT 497
Query: 1587 GF-------WKLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVED 1639
WK +W+ P K++ F + V R+ +P R +A C C + +
Sbjct: 498 SSSAASSFPWKKLWRTPVPNKMKHFAYRVARNIIPCRENLQRRGIAVQNECPTCNHQIPE 677
Query: 1640 A 1640
+
Sbjct: 678 S 680
>AV780445
Length = 504
Score = 41.2 bits (95), Expect(2) = 6e-06
Identities = 27/85 (31%), Positives = 39/85 (45%), Gaps = 7/85 (8%)
Frame = +1
Query: 1683 NTLFC-AMAWNLWKWRNSFVFEQVPWQPGDVMRRIHLDTVEFHRLAQ------GTQRSNP 1735
N++ C A W +WKWRN+ VFE PW + R++ + EF + G+ S+
Sbjct: 211 NSVKCLAGVW*VWKWRNNMVFEDSPWSVDEAWRKLGHEHDEFITFSDGDAGDPGSWLSSR 390
Query: 1736 IASDGINSVMLYTDGSWNPQVNRMG 1760
+V L DGS Q MG
Sbjct: 391 WQPPRQGTVKLNVDGSGREQDRCMG 465
Score = 27.3 bits (59), Expect(2) = 6e-06
Identities = 14/28 (50%), Positives = 16/28 (57%), Gaps = 2/28 (7%)
Frame = +2
Query: 1629 SCIRCGAVVEDAEHVLRRCPGS--VWVR 1654
S RC A ED H LR CP S +W+R
Sbjct: 56 SFFRCPAPEEDI*HCLRDCPHSQEIWLR 139
>BP068806
Length = 418
Score = 44.7 bits (104), Expect = 1e-04
Identities = 24/59 (40%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Frame = +3
Query: 1598 PEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC--PGSVWVR 1654
PEK+R ++W + + A+ N+ R R LAT+ + RC ED H LR C VWVR
Sbjct: 153 PEKIRVWVWFIMQGAIQVNSFRFRCKLATSPAGPRCQHQFEDIWHCLRDCRIARDVWVR 329
>CB826728
Length = 509
Score = 42.7 bits (99), Expect = 5e-04
Identities = 29/98 (29%), Positives = 40/98 (40%), Gaps = 11/98 (11%)
Frame = +1
Query: 1561 WREASDDIYSASSAYQFL---NEDGLPETG--------FWKLIWKASAPEKVRFFLWLVG 1609
W +SD YS+ S Y+F+ N+ LP T WK +W AS+ + + +W
Sbjct: 214 WPGSSDGWYSSKSGYEFIRLENKSLLPSTSPAAAVPSLIWKTVWSASSLPRCKEVMWRAC 393
Query: 1610 RDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC 1647
LP + R L S G E HVL C
Sbjct: 394 AGYLPVRSALKRRGLDVDPSSPWSGLEDEAEVHVLLTC 507
>AV768875
Length = 232
Score = 41.6 bits (96), Expect = 0.001
Identities = 18/39 (46%), Positives = 22/39 (56%)
Frame = +1
Query: 706 GTSQQQRASNMLNMLEACNFIDLGFMGSKFTWERRVNGR 744
G RA +L+ CN IDLG +G KFTW R+ N R
Sbjct: 112 GEFHPNRAQRFATVLDDCNLIDLGMVGGKFTWFRKRNNR 228
>AV412895
Length = 405
Score = 40.8 bits (94), Expect = 0.002
Identities = 29/89 (32%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Frame = +1
Query: 1569 YSASSAYQFLNEDGLPETG-FWKLIWKASAPEKVRFFLW--LVGRDALPTNA-KRHRHHL 1624
+S +S++ FL + L E + IW AP ++ F+W L+GR N KR H
Sbjct: 10 FSVNSSFVFL*DQYLEEPDPVFN*IWMVPAPLNIKAFVWRVLLGRIQTRDNLLKRQVIHN 189
Query: 1625 ATTASCIRCGAVVEDAEHVLRRCPGSVWV 1653
A A C CG E H+L C S+ +
Sbjct: 190 ALDAICPLCGLAEESGSHLLFSCAESMLI 276
>TC14334 weakly similar to UP|AMPL_YEAST (P14904) Vacuolar aminopeptidase I
precursor (Polypeptidase) (Leucine aminopeptidase IV)
(LAPIV) (Aminopeptidase III) (Aminopeptidase yscI) ,
partial (4%)
Length = 1267
Score = 39.7 bits (91), Expect = 0.004
Identities = 29/116 (25%), Positives = 52/116 (44%), Gaps = 7/116 (6%)
Frame = +3
Query: 960 VQVRRNRIMENQTPTIPMEERDKLVA------PVSLEEVRSAIMSMKSFKAPGPDGFHAY 1013
+++ EN+T +EE D+ V++E +++ + + FHA
Sbjct: 420 IRLEEKEKKENETRLQILEEADEYKIGFYRKREVNVENTKASNREREKLFLASGEKFHAE 599
Query: 1014 FYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLI-PKVDNPTKLKDLRPISL 1068
K YW +GE + H V + + G + E +++V+ PK PT L +R I L
Sbjct: 600 ADKNYWKAIGELIPHEVPAIQKRGKKDKEKKPSIIVIQGPKPGKPTDLSRMRQILL 767
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.336 0.145 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,789,911
Number of Sequences: 28460
Number of extensions: 588301
Number of successful extensions: 4601
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 4462
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4590
length of query: 1904
length of database: 4,897,600
effective HSP length: 104
effective length of query: 1800
effective length of database: 1,937,760
effective search space: 3487968000
effective search space used: 3487968000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0228a.4


