

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.2
(184 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
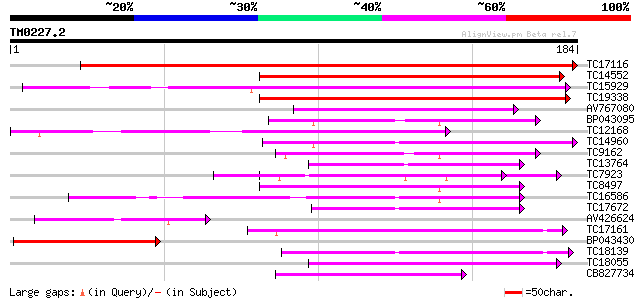
Score E
Sequences producing significant alignments: (bits) Value
TC17116 weakly similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-t... 337 1e-93
TC14552 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, ... 120 1e-28
TC15929 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, ... 119 4e-28
TC19338 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m ... 112 4e-26
AV767080 62 8e-11
BP043095 60 2e-10
TC12168 similar to UP|Q8L7S9 (Q8L7S9) At1g43560/T10P12_4, partia... 60 2e-10
TC14960 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%) 60 2e-10
TC9162 similar to UP|Q9XF70 (Q9XF70) Thioredoxin h, partial (86%) 53 4e-08
TC13764 similar to UP|Q9C9Y6 (Q9C9Y6) Thioredoxin-like protein, ... 52 5e-08
TC7923 similar to UP|PDI_MEDSA (P29828) Protein disulfide isomer... 49 7e-07
TC8497 similar to UP|PDA6_MEDSA (P38661) Probable protein disulf... 49 7e-07
TC16586 weakly similar to UP|Q93VQ9 (Q93VQ9) At1g31020/F17F8_6 (... 47 2e-06
TC17672 similar to UP|Q93X83 (Q93X83) Hsp70 interacting protein/... 46 3e-06
AV426624 43 4e-05
TC17161 homologue to UP|THIF_PEA (P29450) Thioredoxin F-type, ch... 42 8e-05
BP043430 40 2e-04
TC18139 40 2e-04
TC18055 similar to UP|CAE46765 (CAE46765) NADPH thioredoxin redu... 40 3e-04
CB827734 40 3e-04
>TC17116 weakly similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3,
chloroplast precursor (TRX-M3), partial (55%)
Length = 798
Score = 337 bits (863), Expect = 1e-93
Identities = 161/161 (100%), Positives = 161/161 (100%)
Frame = +3
Query: 24 SSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSLRQETRATPIT 83
SSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSLRQETRATPIT
Sbjct: 3 SSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSLRQETRATPIT 182
Query: 84 KDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIAD 143
KDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIAD
Sbjct: 183 KDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIAD 362
Query: 144 DYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLKP 184
DYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLKP
Sbjct: 363 DYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLKP 485
>TC14552 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (55%)
Length = 1016
Score = 120 bits (301), Expect = 1e-28
Identities = 50/99 (50%), Positives = 74/99 (74%)
Frame = +1
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
IT W++ +L+S+ PVLVEF+A WCGPCRM+H +ID++AK+YAG+L C+ +NTD
Sbjct: 310 ITDANWQSLVLESEFPVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDESPST 489
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
A Y I+++P VM+F+DG+K D VIG +PK ++IE+
Sbjct: 490 ATRYGIRSIPTVMIFRDGEKKDTVIGAVPKSTLTSSIEK 606
>TC15929 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (49%)
Length = 1406
Score = 119 bits (297), Expect = 4e-28
Identities = 73/180 (40%), Positives = 102/180 (56%), Gaps = 2/180 (1%)
Frame = +1
Query: 5 STSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHL 64
ST S+T SS S+SSS S SS+R SL + L
Sbjct: 109 STLPSVTVSSPISASSSLSGRR------SSARLLHYTSLKLR-----SVAGTRLTASKFA 255
Query: 65 PRSHKIRSLRQET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAK 122
PR ++ Q T T IT W++ +++SD VLVEF+A WCGPCRM+H VID++AK
Sbjct: 256 PRRGRVVCEAQNTAVEVTSITDANWQSLVIESDTAVLVEFWAPWCGPCRMIHPVIDELAK 435
Query: 123 EYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
EYAG+L C+ +NTD A Y I+++P V++FKDG+K DAVIG++P + +I + L
Sbjct: 436 EYAGKLKCYKLNTDESPSTATRYGIRSIPTVIIFKDGEKKDAVIGSVP*TTVIASIGKFL 615
>TC19338 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (65%)
Length = 476
Score = 112 bits (280), Expect = 4e-26
Identities = 45/101 (44%), Positives = 74/101 (72%)
Frame = +2
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T+ W + ++ S+IPVLV+F+A WCGPCRM+ +ID++AKEYAG++ C+ +NTD I
Sbjct: 41 VTESSWNDLVIASEIPVLVDFWAPWCGPCRMIAPLIDELAKEYAGKIACYKLNTDDSPNI 220
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
A Y I+++P V+ FK+G+K ++VIG +PK ++E+ +
Sbjct: 221 ATQYGIRSIPTVLFFKNGEKKESVIGAVPKSTLTASLEKYI 343
>AV767080
Length = 559
Score = 61.6 bits (148), Expect = 8e-11
Identities = 23/73 (31%), Positives = 44/73 (59%)
Frame = -1
Query: 93 KSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPV 152
K D+P++++FYA+WCGPC ++ + ++ +A EY + V+TD + + A D +++ +P
Sbjct: 532 KXDVPLIIDFYATWCGPCILMAQELEMLAVEYEQNVTIVKVDTDDEYEFARDMQVRGLPT 353
Query: 153 VMLFKDGQKCDAV 165
V DA+
Sbjct: 352 VFFISPDPNKDAI 314
>BP043095
Length = 396
Score = 60.5 bits (145), Expect = 2e-10
Identities = 30/93 (32%), Positives = 50/93 (53%), Gaps = 5/93 (5%)
Frame = +1
Query: 85 DLWENSILKSDIP---VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD--M 139
D W + + K + ++V+F ASWCGPCR + + ++AK+Y N + D D
Sbjct: 103 DAWTDHLNKGNESKKLIVVDFTASWCGPCRFIAPYLGELAKKYT---NVIFLKVDVDELK 273
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKE 172
+A D+ ++A+P M K+G D V+G +E
Sbjct: 274 SVAQDWAVEAMPTFMFVKEGSIVDKVVGAKKEE 372
>TC12168 similar to UP|Q8L7S9 (Q8L7S9) At1g43560/T10P12_4, partial (50%)
Length = 408
Score = 60.5 bits (145), Expect = 2e-10
Identities = 43/144 (29%), Positives = 68/144 (46%), Gaps = 1/144 (0%)
Frame = +1
Query: 1 MASISTSI-SLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQ 59
MA S+ I SL S+ SS S SSS+ L S PSL FP R+
Sbjct: 31 MAVTSSVIPSLNSTRLGSSPSPSSSTRL---------SSTTPSLHFPSRTRVLRIGTSRA 183
Query: 60 NPPHLPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDD 119
P LP +A T +++ + SD PV V+FYA+WCGPC+ + ++D+
Sbjct: 184 PPRFLP----------VVQAKKQTFSSFDDMLANSDKPVFVDFYATWCGPCQFMVPILDE 333
Query: 120 IAKEYAGRLNCFLVNTDTDMQIAD 143
++ ++ ++T+ +IA+
Sbjct: 334 VSTRLKDKIQVVKIDTEKYPEIAN 405
>TC14960 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%)
Length = 688
Score = 60.5 bits (145), Expect = 2e-10
Identities = 28/105 (26%), Positives = 57/105 (53%), Gaps = 3/105 (2%)
Frame = +1
Query: 83 TKDLWENSILKSDIP---VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDM 139
T + W+ + K ++ ++V+F ASWCGPCR + ++ +IAK+ + V+ D
Sbjct: 115 TVEAWKEHLEKGNVSKKLIVVDFTASWCGPCRFIAPILAEIAKKLP-HVVFLKVDVDELK 291
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLKP 184
+A+++ ++A+P + K+G+ D V+G +E + + P
Sbjct: 292 TVAEEWNVEAMPTFLFLKEGKAVDKVVGAKKEELQLTITKHAEAP 426
>TC9162 similar to UP|Q9XF70 (Q9XF70) Thioredoxin h, partial (86%)
Length = 660
Score = 52.8 bits (125), Expect = 4e-08
Identities = 30/91 (32%), Positives = 45/91 (48%), Gaps = 5/91 (5%)
Frame = +3
Query: 87 WE---NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD--MQI 141
WE N + P++V F ASWC P ++ +D+A Y L +N D D +
Sbjct: 96 WEFYVNQASNQNCPIVVHFSASWCMPSVAMNPFFEDMASSYPDFL---FLNVDVDEVKDV 266
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKE 172
A EIKA+P + KDG + ++G P+E
Sbjct: 267 ATRLEIKAMPTFVFLKDGAPLEKLVGANPEE 359
>TC13764 similar to UP|Q9C9Y6 (Q9C9Y6) Thioredoxin-like protein, partial
(73%)
Length = 585
Score = 52.4 bits (124), Expect = 5e-08
Identities = 22/70 (31%), Positives = 40/70 (56%)
Frame = +3
Query: 98 VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFK 157
V+ F A+WCGPC+++ +++++Y + LV+ D + ++IKA P K
Sbjct: 39 VIANFSATWCGPCKIIAPYYCELSEKYTSMM-FLLVDVDELTDFSTSWDIKATPTFFFLK 215
Query: 158 DGQKCDAVIG 167
DGQ+ D ++G
Sbjct: 216 DGQQLDQLVG 245
>TC7923 similar to UP|PDI_MEDSA (P29828) Protein disulfide isomerase
precursor (PDI) , partial (89%)
Length = 1861
Score = 48.5 bits (114), Expect = 7e-07
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 2/100 (2%)
Frame = +2
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQ- 140
+ D ++ + KS VL+EFYA WCG C+ + ++D++A Y + + D
Sbjct: 1190 VVGDSLQDVVFKSGKNVLLEFYAPWCGHCKSLAPILDEVAVSYQNDADIVIAKFDATAND 1369
Query: 141 -IADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIE 179
+ +E++ P + K G KE + IE
Sbjct: 1370 VPTESFEVQGYPTLYFISSSGKISPYDGGRTKEDIIQFIE 1489
Score = 40.4 bits (93), Expect = 2e-04
Identities = 25/102 (24%), Positives = 47/102 (45%), Gaps = 7/102 (6%)
Frame = +2
Query: 67 SHKIRSLRQETRATPITKDL--WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEY 124
S +I ++ E + +T D + + K D ++VEFYA WCG C+ + + A
Sbjct: 107 SSQIHAIESEAKEFVLTLDNSNFSEIVSKHDF-IVVEFYAPWCGHCKNLAPEYEKAAAVL 283
Query: 125 AGR-----LNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQK 161
+ L N + + +A YE++ P + + ++G K
Sbjct: 284 SSHDPPIVLAKVDANEENNKDLASQYEVRGFPTIKILRNGGK 409
>TC8497 similar to UP|PDA6_MEDSA (P38661) Probable protein disulfide
isomerase A6 precursor (P5) , partial (95%)
Length = 1533
Score = 48.5 bits (114), Expect = 7e-07
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 2/88 (2%)
Frame = +2
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD--M 139
+T + + +L VLVEFYA WCG C+ + + +A + + + N D D
Sbjct: 560 LTAENFNEVVLDETKDVLVEFYAPWCGHCKSLAPTYEKVAAAFKLDGDVVIANLDADKYR 739
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIG 167
+A+ YE+ P + F G K G
Sbjct: 740 DLAEKYEVSGFPTLKFFPKGNKAGEEYG 823
Score = 36.2 bits (82), Expect = 0.003
Identities = 19/80 (23%), Positives = 34/80 (41%), Gaps = 2/80 (2%)
Frame = +2
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD--M 139
+++D +E + D LVEFYA WCG C+ + + + + + + D D
Sbjct: 206 LSEDNFEKEV-GQDKGALVEFYAPWCGHCKKLAPEYEKLGGSFKKAKSVLIAKVDCDEHK 382
Query: 140 QIADDYEIKAVPVVMLFKDG 159
+ Y + P + F G
Sbjct: 383 SVCSKYGVSGYPTLQWFPKG 442
>TC16586 weakly similar to UP|Q93VQ9 (Q93VQ9) At1g31020/F17F8_6 (Thioredoxin
o) (At2g20920/F5H14.11), partial (35%)
Length = 898
Score = 47.0 bits (110), Expect = 2e-06
Identities = 35/150 (23%), Positives = 68/150 (45%), Gaps = 2/150 (1%)
Frame = +2
Query: 20 SSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSLRQETRA 79
SS S+S F ++ S L P PFH H ++ P + + + E
Sbjct: 170 SSLSNSKTTSLFAATIASTLLP----PFH--------HSRSLCSAPANPGVVLVNSEEEF 313
Query: 80 TPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD- 138
I + +NS+ ++ F A WCGPC+ + ++ +++K+Y + + ++ D +
Sbjct: 314 NRILTKIQDNSL-----HAIIYFTAVWCGPCKFISPIVGELSKKYP-HVTTYKIDMDQEA 475
Query: 139 -MQIADDYEIKAVPVVMLFKDGQKCDAVIG 167
+I +VP + F +G+K D ++G
Sbjct: 476 LRGTLTKLQISSVPTLHFFLNGKKADELVG 565
>TC17672 similar to UP|Q93X83 (Q93X83) Hsp70 interacting protein/thioredoxin
chimera, partial (33%)
Length = 709
Score = 46.2 bits (108), Expect = 3e-06
Identities = 20/69 (28%), Positives = 39/69 (55%)
Frame = +1
Query: 99 LVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKD 158
++ F A+WCGPCR + + +A++Y ++ V+ D +A + I +VP K+
Sbjct: 163 ILYFTATWCGPCRFISPIYTSLAEKYP-KVVFLKVDIDEARDVAARWNISSVPSFFFVKN 339
Query: 159 GQKCDAVIG 167
G++ D+ +G
Sbjct: 340 GKEVDSAVG 366
>AV426624
Length = 422
Score = 42.7 bits (99), Expect = 4e-05
Identities = 30/59 (50%), Positives = 35/59 (58%), Gaps = 2/59 (3%)
Frame = -2
Query: 9 SLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLR--ITHKPLHLQNPPHLP 65
S +SSSSSSSSSSSSSS L +PF SS SHL F+LR +TH +P P
Sbjct: 400 SSSSSSSSSSSSSSSSSSL*EPFYSS--SHLKKQKPQIFNLRTLMTHTQTRYSHPVRFP 230
>TC17161 homologue to UP|THIF_PEA (P29450) Thioredoxin F-type, chloroplast
precursor (TRX-F), partial (72%)
Length = 850
Score = 41.6 bits (96), Expect = 8e-05
Identities = 26/105 (24%), Positives = 47/105 (44%), Gaps = 1/105 (0%)
Frame = +2
Query: 78 RATPITKD-LWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTD 136
+ T + KD W D V+++ + WCGPC+++ ++A++ + L
Sbjct: 254 KVTEVNKDTFWPIVNAAGDKTVVLDMFTQWCGPCKVIAPKYQELAEKNLDVVFLKLDCNQ 433
Query: 137 TDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
+ +A + IK VP + KD + + G + V AIE V
Sbjct: 434 DNKPLAKELGIKVVPTFKILKDSKIVKEITGAKYDDL-VAAIETV 565
>BP043430
Length = 493
Score = 40.0 bits (92), Expect = 2e-04
Identities = 27/48 (56%), Positives = 32/48 (66%)
Frame = +3
Query: 2 ASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHL 49
+S+++S+SL SSSS SSSSSSSS F SSS S PSLS P L
Sbjct: 249 SSLNSSLSLPSSSSLLSSSSSSSSSTSLSFPSSSLSLPLPSLSSPLAL 392
Score = 28.1 bits (61), Expect = 0.94
Identities = 21/40 (52%), Positives = 25/40 (62%)
Frame = +3
Query: 7 SISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFP 46
S+S +SS +SS S SSSS L SSS S + SLSFP
Sbjct: 234 SLSSSSSLNSSLSLPSSSSLLS----SSSSSSSSTSLSFP 341
>TC18139
Length = 429
Score = 40.0 bits (92), Expect = 2e-04
Identities = 20/95 (21%), Positives = 44/95 (46%)
Frame = +1
Query: 89 NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIK 148
+ ++ V + F+ASWC + + ++ ++ ++ + V + +I++ Y +
Sbjct: 70 DEVVHGGAAVALHFWASWCEASKHMDKIFSHLSTDFP-HAHFLRVEAEEQPEISEAYSVS 246
Query: 149 AVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
AVP + KDG+ D G P N + +V +
Sbjct: 247 AVPFFVFCKDGKTVDTSEGADPSSL-ANKVAKVAR 348
>TC18055 similar to UP|CAE46765 (CAE46765) NADPH thioredoxin reductase,
partial (20%)
Length = 748
Score = 39.7 bits (91), Expect = 3e-04
Identities = 20/82 (24%), Positives = 37/82 (44%)
Frame = +1
Query: 98 VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFK 157
+ V + + CGPCR + ++ + E+ ++ ++ + D ++A I P V FK
Sbjct: 259 ICVLYTSPTCGPCRTLKPILSKVIDEFDQNVHFVEIDIEEDQEVAAAAGIMGTPCVQFFK 438
Query: 158 DGQKCDAVIGTMPKEFYVNAIE 179
+ + V G K Y IE
Sbjct: 439 NKEMLKTVSGVKMKREYREFIE 504
>CB827734
Length = 556
Score = 39.7 bits (91), Expect = 3e-04
Identities = 18/62 (29%), Positives = 33/62 (53%)
Frame = +1
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
+++ +L S VLVEF+A WCG C+ + + + A G + ++ D +A +Y
Sbjct: 160 FKSKVLNSKGVVLVEFFAPWCGHCKALTPIWEKAATVLKGVVTVAALDADAHQALAQEYG 339
Query: 147 IK 148
I+
Sbjct: 340 IR 345
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,208,828
Number of Sequences: 28460
Number of extensions: 98031
Number of successful extensions: 5113
Number of sequences better than 10.0: 431
Number of HSP's better than 10.0 without gapping: 1860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3270
length of query: 184
length of database: 4,897,600
effective HSP length: 85
effective length of query: 99
effective length of database: 2,478,500
effective search space: 245371500
effective search space used: 245371500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0227.2


