

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
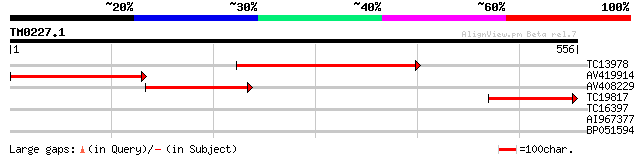
Query= TM0227.1
(556 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13978 similar to UP|MCCB_ARATH (Q9LDD8) Methylcrotonyl-CoA car... 363 e-101
AV419914 274 3e-74
AV408229 205 1e-53
TC19817 weakly similar to UP|MCCB_ARATH (Q9LDD8) Methylcrotonyl-... 166 1e-41
TC16397 UP|ACCD_LOTJA (Q9BBS1) Acetyl-coenzyme A carboxylase car... 32 0.35
AI967377 28 5.1
BP051594 27 6.6
>TC13978 similar to UP|MCCB_ARATH (Q9LDD8) Methylcrotonyl-CoA carboxylase
beta chain, mitochondrial precursor
(3-Methylcrotonyl-CoA carboxylase 2) (MCCase beta
subunit) (3-methylcrotonyl-CoA:carbon dioxide ligase
beta subunit) , partial (29%)
Length = 545
Score = 363 bits (931), Expect = e-101
Identities = 181/181 (100%), Positives = 181/181 (100%)
Frame = +1
Query: 223 NGTIFLAGPPLVKAATGEEVSAEDLGGATVHCKTSGVSDYFAQDEFHALALGRNIIKNLH 282
NGTIFLAGPPLVKAATGEEVSAEDLGGATVHCKTSGVSDYFAQDEFHALALGRNIIKNLH
Sbjct: 1 NGTIFLAGPPLVKAATGEEVSAEDLGGATVHCKTSGVSDYFAQDEFHALALGRNIIKNLH 180
Query: 283 MAGKDVLANGLQNLSYEYKEPLYDVNELRSIAPTDLKKQFDVRSVIARIVDGSEFDEFKK 342
MAGKDVLANGLQNLSYEYKEPLYDVNELRSIAPTDLKKQFDVRSVIARIVDGSEFDEFKK
Sbjct: 181 MAGKDVLANGLQNLSYEYKEPLYDVNELRSIAPTDLKKQFDVRSVIARIVDGSEFDEFKK 360
Query: 343 LYGTTLVTGFARIYGQPVGIIGNNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMV 402
LYGTTLVTGFARIYGQPVGIIGNNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMV
Sbjct: 361 LYGTTLVTGFARIYGQPVGIIGNNGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMV 540
Query: 403 G 403
G
Sbjct: 541 G 543
>AV419914
Length = 422
Score = 274 bits (700), Expect = 3e-74
Identities = 134/134 (100%), Positives = 134/134 (100%)
Frame = +2
Query: 1 MIGLIGRKASWASSNLVGRRGFCLGILPHSNGGATAMEDLLSQLQSHVHKALAGGGAEAV 60
MIGLIGRKASWASSNLVGRRGFCLGILPHSNGGATAMEDLLSQLQSHVHKALAGGGAEAV
Sbjct: 20 MIGLIGRKASWASSNLVGRRGFCLGILPHSNGGATAMEDLLSQLQSHVHKALAGGGAEAV 199
Query: 61 KRNTSRNKLLPRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGVVTGIGPVHGRLCMF 120
KRNTSRNKLLPRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGVVTGIGPVHGRLCMF
Sbjct: 200 KRNTSRNKLLPRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGVVTGIGPVHGRLCMF 379
Query: 121 VANDPTVKGGTYYP 134
VANDPTVKGGTYYP
Sbjct: 380 VANDPTVKGGTYYP 421
>AV408229
Length = 402
Score = 205 bits (522), Expect = 1e-53
Identities = 101/105 (96%), Positives = 103/105 (97%)
Frame = +3
Query: 134 PITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIFYNQALMSAEGI 193
PITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIFYNQALMSAE I
Sbjct: 3 PITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIFYNQALMSAERI 182
Query: 194 PQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLVKAAT 238
PQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLVK ++
Sbjct: 183 PQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLVKVSS 317
>TC19817 weakly similar to UP|MCCB_ARATH (Q9LDD8) Methylcrotonyl-CoA
carboxylase beta chain, mitochondrial precursor
(3-Methylcrotonyl-CoA carboxylase 2) (MCCase beta
subunit) (3-methylcrotonyl-CoA:carbon dioxide ligase
beta subunit) , partial (15%)
Length = 446
Score = 166 bits (419), Expect = 1e-41
Identities = 82/87 (94%), Positives = 86/87 (98%)
Frame = +3
Query: 470 QAAGVLSQIEKTNKKKQGIQWSKEEEEKFKTKVVEAYEVEGSPYYSTARLWDDGIIDPAD 529
QAAGVLSQIEKT++KK GIQWS+EEEEKF+TKVVEAYEVEGSPYYSTARLWDDGIIDPAD
Sbjct: 6 QAAGVLSQIEKTHQKKPGIQWSQEEEEKFQTKVVEAYEVEGSPYYSTARLWDDGIIDPAD 185
Query: 530 TRKVIGLSISASLNRAIEDTKFGVFRM 556
TRKVIGLSISASLNRAIEDTKFGVFRM
Sbjct: 186 TRKVIGLSISASLNRAIEDTKFGVFRM 266
>TC16397 UP|ACCD_LOTJA (Q9BBS1) Acetyl-coenzyme A carboxylase carboxyl
transferase subunit beta (ACCASE beta chain) , complete
Length = 1514
Score = 31.6 bits (70), Expect = 0.35
Identities = 38/192 (19%), Positives = 76/192 (38%), Gaps = 17/192 (8%)
Frame = +3
Query: 68 KLLPRERIDRLLDPGS------SFLELSQLAGH---ELYEEPLPS--------GGVVTGI 110
K+ +RI+ +DPG+ + + + H E Y++ + S V TG
Sbjct: 792 KMSSSDRIELSIDPGTWNPMDEDMISVDPIEFHSEEEPYKDRIDSYQKTTGLTEAVQTGT 971
Query: 111 GPVHGRLCMFVANDPTVKGGTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQAD 170
G ++G D GG+ + +K R E A+ LP + + SGGA + +++
Sbjct: 972 GHLNGIPVAIGIMDFEFMGGSMGSVVGEKITRLVEYATNQLLPLIVVCASGGARMQERSL 1151
Query: 171 VFPDRENFGRIFYNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAG 230
Y+ L + + ++++ T G M + ++ + N I AG
Sbjct: 1152SLMQMAKISSALYDYQL--NKKLFYVSILTSPTTGGVTASFGMLGDIIIAEPNAYIAFAG 1325
Query: 231 PPLVKAATGEEV 242
+++ + V
Sbjct: 1326KRVIEQTLNKAV 1361
>AI967377
Length = 436
Score = 27.7 bits (60), Expect = 5.1
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = -1
Query: 373 SALKGAHFIEICTQRNIPLVFLQNITGFMVGSRS 406
S L +HF I + N+P+ FL N+TG M R+
Sbjct: 376 SVLSRSHFQIIYDK*NLPISFLFNLTGCMTQPRN 275
>BP051594
Length = 376
Score = 27.3 bits (59), Expect = 6.6
Identities = 10/28 (35%), Positives = 18/28 (63%)
Frame = +2
Query: 77 RLLDPGSSFLELSQLAGHELYEEPLPSG 104
R+++PGS + + L GH L++ +P G
Sbjct: 218 RVIEPGSLHVTRASLTGHHLFQVGIPLG 301
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,519,115
Number of Sequences: 28460
Number of extensions: 108969
Number of successful extensions: 475
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 474
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 475
length of query: 556
length of database: 4,897,600
effective HSP length: 95
effective length of query: 461
effective length of database: 2,193,900
effective search space: 1011387900
effective search space used: 1011387900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0227.1


