

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0221.10
(296 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
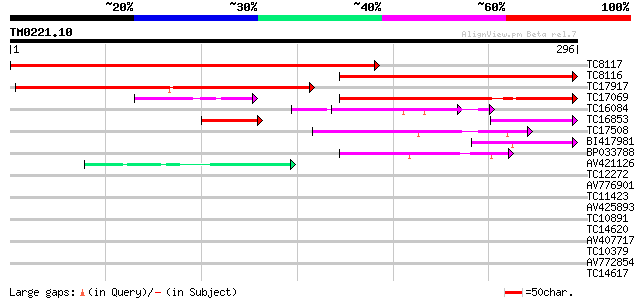
Score E
Sequences producing significant alignments: (bits) Value
TC8117 similar to UP|Q9XGN6 (Q9XGN6) ESTs C99174(E10437), partia... 412 e-116
TC8116 similar to UP|Q9XGN6 (Q9XGN6) ESTs C99174(E10437), partia... 257 1e-69
TC17917 similar to GB|AAO63324.1|28950801|BT005260 At5g06770 {Ar... 221 1e-58
TC17069 similar to UP|Q9XGN6 (Q9XGN6) ESTs C99174(E10437), parti... 164 2e-41
TC16084 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H... 47 3e-06
TC16853 similar to UP|Q93ZP3 (Q93ZP3) At1g32359/F27G20.10, parti... 47 5e-06
TC17508 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H... 44 2e-05
BI417981 41 3e-04
BP033788 41 3e-04
AV421126 39 8e-04
TC12272 35 0.012
AV776901 35 0.012
TC11423 35 0.015
AV425893 35 0.020
TC10891 similar to UP|Q941Q3 (Q941Q3) Floral homeotic protein HU... 33 0.044
TC14620 33 0.075
AV407717 33 0.075
TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast ... 32 0.098
AV772854 32 0.098
TC14617 similar to UP|Q42361 (Q42361) Transcribed sequence 1087 ... 32 0.13
>TC8117 similar to UP|Q9XGN6 (Q9XGN6) ESTs C99174(E10437), partial (41%)
Length = 766
Score = 412 bits (1058), Expect = e-116
Identities = 193/193 (100%), Positives = 193/193 (100%)
Frame = +3
Query: 1 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 60
MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY
Sbjct: 186 MDFRKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHY 365
Query: 61 VPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKC 120
VPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKC
Sbjct: 366 VPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKC 545
Query: 121 HFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLA 180
HFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLA
Sbjct: 546 HFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLA 725
Query: 181 GAIIGKGGVNSKQ 193
GAIIGKGGVNSKQ
Sbjct: 726 GAIIGKGGVNSKQ 764
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/32 (56%), Positives = 23/32 (71%), Gaps = 2/32 (6%)
Frame = +3
Query: 267 KTKLCENF--AKGTCTFGERCHFAHGPAELRK 296
KT++C F A+G C FG++CHFAHG EL K
Sbjct: 486 KTRICNKFNTAEG-CKFGDKCHFAHGEWELGK 578
Score = 28.5 bits (62), Expect = 1.4
Identities = 12/24 (50%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Frame = +3
Query: 267 KTKLCENF-AKGTCTFGERCHFAH 289
K+K C F + C FGE CHF H
Sbjct: 291 KSKPCTKFFSTAGCPFGESCHFLH 362
>TC8116 similar to UP|Q9XGN6 (Q9XGN6) ESTs C99174(E10437), partial (31%)
Length = 595
Score = 257 bits (657), Expect = 1e-69
Identities = 124/124 (100%), Positives = 124/124 (100%)
Frame = +2
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV 232
ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV
Sbjct: 2 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV 181
Query: 233 KDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPA 292
KDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPA
Sbjct: 182 KDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPA 361
Query: 293 ELRK 296
ELRK
Sbjct: 362 ELRK 373
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/32 (56%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Frame = +2
Query: 101 KTRICNKFNTAEG-CKFGDKCHFAHGEWELGK 131
KT++C F A+G C FG++CHFAHG EL K
Sbjct: 284 KTKLCENF--AKGTCTFGERCHFAHGPAELRK 373
Score = 28.5 bits (62), Expect = 1.4
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = +2
Query: 36 KSKPCTKFFSTAGCPFGESCHFLH 59
K+K C F + C FGE CHF H
Sbjct: 284 KTKLCENF-AKGTCTFGERCHFAH 352
>TC17917 similar to GB|AAO63324.1|28950801|BT005260 At5g06770 {Arabidopsis
thaliana;}, partial (35%)
Length = 587
Score = 221 bits (563), Expect = 1e-58
Identities = 100/159 (62%), Positives = 123/159 (76%), Gaps = 3/159 (1%)
Frame = +1
Query: 4 RKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPG 63
RKRGRPE FN NGG KKSK E+ES ++G+GSKSKPCTKFFST+GCPFG+ CHFLHYVPG
Sbjct: 100 RKRGRPEGAFNGNGGAKKSKPEMESFTTGLGSKSKPCTKFFSTSGCPFGDGCHFLHYVPG 279
Query: 64 GYNAVAHMMNLAPSAQAPP--RNVAAPPPPVPNGST-PAVKTRICNKFNTAEGCKFGDKC 120
G+ A++ M+N+ S P RN PPP P+GS+ P VKTR+CNK+N++EGCKFGD+C
Sbjct: 280 GFKAISQMINVGSSPTIPQMGRN-PVPPPSFPDGSSPPVVKTRMCNKYNSSEGCKFGDRC 456
Query: 121 HFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPP 159
HFAHGEWELG+ P ++D R G + R GGR+EPPP
Sbjct: 457 HFAHGEWELGRPTLPPYEDPRGFGQMQSNRGGGRIEPPP 573
Score = 37.0 bits (84), Expect = 0.004
Identities = 15/29 (51%), Positives = 19/29 (64%), Gaps = 1/29 (3%)
Frame = +1
Query: 267 KTKLCENFAKGT-CTFGERCHFAHGPAEL 294
KT++C + C FG+RCHFAHG EL
Sbjct: 397 KTRMCNKYNSSEGCKFGDRCHFAHGEWEL 483
Score = 26.9 bits (58), Expect = 4.1
Identities = 11/24 (45%), Positives = 14/24 (57%), Gaps = 1/24 (4%)
Frame = +1
Query: 267 KTKLCENF-AKGTCTFGERCHFAH 289
K+K C F + C FG+ CHF H
Sbjct: 196 KSKPCTKFFSTSGCPFGDGCHFLH 267
>TC17069 similar to UP|Q9XGN6 (Q9XGN6) ESTs C99174(E10437), partial (33%)
Length = 577
Score = 164 bits (415), Expect = 2e-41
Identities = 86/125 (68%), Positives = 98/125 (77%), Gaps = 1/125 (0%)
Frame = +2
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV 232
IS+ ASLAGAIIGK GVNSKQICR TGAKLSIREH+SDPNL+NIELEGSFDQI +AS MV
Sbjct: 2 ISINASLAGAIIGKNGVNSKQICRVTGAKLSIREHDSDPNLKNIELEGSFDQINQASAMV 181
Query: 233 KDLLLTL-QMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGP 291
+L+L++ +S PP N H GS N KTKLCENFA+G+CTFGERCHFAHG
Sbjct: 182 HELILSVSSVSGPPAKNFSSQ-----HSAPGS-NLKTKLCENFAQGSCTFGERCHFAHGT 343
Query: 292 AELRK 296
ELR+
Sbjct: 344 DELRR 358
Score = 39.7 bits (91), Expect = 6e-04
Identities = 24/66 (36%), Positives = 37/66 (55%), Gaps = 2/66 (3%)
Frame = +2
Query: 66 NAVAHMMNLAPSA-QAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEG-CKFGDKCHFA 123
+A+ H + L+ S+ PP + P + +KT++C F A+G C FG++CHFA
Sbjct: 170 SAMVHELILSVSSVSGPPAKNFSSQHSAPGSN---LKTKLCENF--AQGSCTFGERCHFA 334
Query: 124 HGEWEL 129
HG EL
Sbjct: 335 HGTDEL 352
Score = 30.0 bits (66), Expect = 0.49
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +2
Query: 32 GVGSKSKPCTKFFSTAGCPFGESCHFLH 59
G K+K C F + C FGE CHF H
Sbjct: 257 GSNLKTKLCENF-AQGSCTFGERCHFAH 337
>TC16084 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;}, partial (29%)
Length = 889
Score = 47.4 bits (111), Expect = 3e-06
Identities = 28/88 (31%), Positives = 50/88 (56%), Gaps = 3/88 (3%)
Frame = +1
Query: 169 ATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNLRN--IELEGSFDQI 225
++ KI + + G IIGKGG K + ++GAK+ + R+ ++DPN N +EL GS + I
Sbjct: 313 SSKKIDIPSGRVGVIIGKGGETIKSLQLRSGAKIQVTRDMDADPNSPNRLVELMGSPEAI 492
Query: 226 KEASNMVKDLLLTLQMSAPPKSNQGGAG 253
+A ++ ++L ++ GG+G
Sbjct: 493 ADAEKLINEVL--------AEAESGGSG 552
Score = 39.3 bits (90), Expect = 8e-04
Identities = 26/93 (27%), Positives = 47/93 (49%), Gaps = 4/93 (4%)
Frame = +1
Query: 148 AGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI--- 204
+G IG R+ G A F + + + G IIGKGG K + ++GA++ +
Sbjct: 547 SGMIGRRLTGQSGSADEFS------MQIPNNKVGLIIGKGGETIKSMQAESGARIQVIPL 708
Query: 205 REHESDPNL-RNIELEGSFDQIKEASNMVKDLL 236
DP+ R ++++GS +QI+ A +V ++
Sbjct: 709 HLPPGDPSTERTLKIDGSPEQIESAKQLVNQII 807
>TC16853 similar to UP|Q93ZP3 (Q93ZP3) At1g32359/F27G20.10, partial (20%)
Length = 722
Score = 46.6 bits (109), Expect = 5e-06
Identities = 19/32 (59%), Positives = 23/32 (71%)
Frame = +1
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKH 132
KTRICNK+ C FG+KCHFAHG EL ++
Sbjct: 58 KTRICNKWEMTGYCPFGNKCHFAHGATELHRY 153
Score = 42.0 bits (97), Expect = 1e-04
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Frame = +1
Query: 252 AGGPGGHGHHGSNNFKTKLCENFAK-GTCTFGERCHFAHGPAELRK 296
A G + +N+KT++C + G C FG +CHFAHG EL +
Sbjct: 13 ASSAGNGANVKPSNWKTRICNKWEMTGYCPFGNKCHFAHGATELHR 150
Score = 34.7 bits (78), Expect = 0.020
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 7/37 (18%)
Frame = +1
Query: 30 SSGVGSKSKP-------CTKFFSTAGCPFGESCHFLH 59
S+G G+ KP C K+ T CPFG CHF H
Sbjct: 19 SAGNGANVKPSNWKTRICNKWEMTGYCPFGNKCHFAH 129
>TC17508 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;}, partial (34%)
Length = 999
Score = 44.3 bits (103), Expect = 2e-05
Identities = 35/121 (28%), Positives = 58/121 (47%), Gaps = 6/121 (4%)
Frame = +1
Query: 159 PGPATGFG-ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LR 214
P A +G + KI + G IIGK G K + Q+GAK+ + R+ ++DPN R
Sbjct: 352 PSFAPSYGHQGGSKKIDIPNGRVGVIIGKSGETIKYLQTQSGAKIQVTRDMDADPNSSTR 531
Query: 215 NIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGH--GHHGSNNFKTKLCE 272
+EL G+ + I A ++ ++L ++ GG+G G G++ F K+
Sbjct: 532 AVELMGTPEAIAHAEKLINEVL--------TEAESGGSGIVTRRIPGQAGADEFVMKVPN 687
Query: 273 N 273
N
Sbjct: 688 N 690
Score = 36.2 bits (82), Expect = 0.007
Identities = 19/68 (27%), Positives = 36/68 (52%), Gaps = 4/68 (5%)
Frame = +1
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP----NLRNIELEGSFDQIKEA 228
+ V + G +IGKGG K + + TGA++ + P R +++EG+ +QI+ A
Sbjct: 673 MKVPNNKVGLVIGKGGETIKSMQQSTGARIQVIPLHLPPGDTSTERTLKIEGNSEQIESA 852
Query: 229 SNMVKDLL 236
+V ++
Sbjct: 853 KQLVNSVI 876
>BI417981
Length = 477
Score = 40.8 bits (94), Expect = 3e-04
Identities = 22/61 (36%), Positives = 26/61 (42%), Gaps = 6/61 (9%)
Frame = +3
Query: 242 SAPPKSNQGGAGGPGGHGHH------GSNNFKTKLCENFAKGTCTFGERCHFAHGPAELR 295
S PP G+ G FKTKLC F GTC + C+FAH ELR
Sbjct: 138 SEPPSKKARGSSATSSSASQERSKAIGKMFFKTKLCCKFRAGTCPYVTNCNFAHSIEELR 317
Query: 296 K 296
+
Sbjct: 318 R 320
Score = 28.9 bits (63), Expect = 1.1
Identities = 16/60 (26%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Frame = +3
Query: 5 KRGRPEPGFNSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFGESCHFLHYV 61
+ G P + G S + S +G K+K C KF CP+ +C+F H +
Sbjct: 129 RSGSEPPSKKARGSSATSSSASQERSKAIGKMFFKTKLCCKF-RAGTCPYVTNCNFAHSI 305
>BP033788
Length = 518
Score = 40.8 bits (94), Expect = 3e-04
Identities = 31/106 (29%), Positives = 50/106 (46%), Gaps = 15/106 (14%)
Frame = +3
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLS-IREH---ESDPNLRNIELEGSFDQIKEA 228
++V G IIG+GG K + ++GA++ I +H D R +++ G QI+ A
Sbjct: 18 VTVPNEKVGLIIGRGGETIKSLQTKSGARIQLIPQHLPEGDDSKERTVQVTGDKRQIEVA 197
Query: 229 SNMVKDLLLTLQMSAPPKSNQG-----------GAGGPGGHGHHGS 263
M+K++ MS P + + G G+GGP G GS
Sbjct: 198 QEMIKEV-----MSQPVRPSTGGYGQQAYRPPRGSGGPPQWGQRGS 320
>AV421126
Length = 416
Score = 39.3 bits (90), Expect = 8e-04
Identities = 30/110 (27%), Positives = 38/110 (34%)
Frame = +2
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
C + T C FG+SC + H P A +NL+P P APP
Sbjct: 140 CHYYMKTGECKFGQSCRYHH--PPDMGASKANVNLSPVGL--PLRPGAPP---------- 277
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAG 149
C + CKFG C F H L + S P+ P G
Sbjct: 278 -----CTHYTQRGICKFGSACKFDHPIGSLSYSPSASSLADMPVAPYPVG 412
Score = 28.9 bits (63), Expect = 1.1
Identities = 18/73 (24%), Positives = 29/73 (39%)
Frame = +2
Query: 18 GIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPS 77
G K+ L + + + PCT + C FG +C F H + ++ +PS
Sbjct: 212 GASKANVNLSPVGLPLRPGAPPCTHYTQRGICKFGSACKFDHPIGS--------LSYSPS 367
Query: 78 AQAPPRNVAAPPP 90
A + AP P
Sbjct: 368 ASSLADMPVAPYP 406
>TC12272
Length = 627
Score = 35.4 bits (80), Expect = 0.012
Identities = 16/31 (51%), Positives = 20/31 (63%)
Frame = +1
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
FKT+LC + +G C C FAHG AELR+
Sbjct: 280 FKTRLCVLYQRGRCN-RHNCSFAHGNAELRR 369
>AV776901
Length = 588
Score = 35.4 bits (80), Expect = 0.012
Identities = 16/31 (51%), Positives = 20/31 (63%)
Frame = +1
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
FKT+LC + +G C C FAHG AELR+
Sbjct: 259 FKTRLCVLYQRGRCN-RHNCSFAHGNAELRR 348
>TC11423
Length = 563
Score = 35.0 bits (79), Expect = 0.015
Identities = 21/90 (23%), Positives = 37/90 (40%)
Frame = +1
Query: 30 SSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPP 89
++ + S + PC+ F + G FL +A H++ PS +P + AP
Sbjct: 100 TAAIPSFAPPCSSFILSTGLL--PRSRFLPLNRKSTSAPKHIIRCKPSPSSPQASFIAPS 273
Query: 90 PPVPNGSTPAVKTRICNKFNTAEGCKFGDK 119
P P T + + + + C FGD+
Sbjct: 274 PQAPFDLTKITSSAVASTSAASRKCVFGDE 363
>AV425893
Length = 414
Score = 34.7 bits (78), Expect = 0.020
Identities = 26/65 (40%), Positives = 31/65 (47%), Gaps = 12/65 (18%)
Frame = +2
Query: 39 PCT--KFFSTAGCPFGESCHFL--------HYVPGGYNAVAHMMNLAPSAQAPPR--NVA 86
PC FFS + SC FL YVPG AVA ++ AP PPR +V+
Sbjct: 131 PCLGPDFFSVSALRVALSCPFLA*RGR*PLFYVPG---AVALLVYFAPLLSPPPRQPHVS 301
Query: 87 APPPP 91
PPPP
Sbjct: 302 RPPPP 316
>TC10891 similar to UP|Q941Q3 (Q941Q3) Floral homeotic protein HUA1, partial
(15%)
Length = 581
Score = 33.5 bits (75), Expect = 0.044
Identities = 23/85 (27%), Positives = 30/85 (35%)
Frame = +3
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
C + C FGE C F H + + S QA + V P +P A
Sbjct: 111 CDFYMKNGECKFGERCKFHHPIDRSAPLL--------SKQASQQTVKLTPAGLPRREGAA 266
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAH 124
+C + CKFG C F H
Sbjct: 267 ----MCPYYLKTGTCKFGATCKFDH 329
Score = 26.9 bits (58), Expect = 4.1
Identities = 10/24 (41%), Positives = 12/24 (49%)
Frame = +3
Query: 40 CTKFFSTAGCPFGESCHFLHYVPG 63
C + T C FG +C F H PG
Sbjct: 270 CPYYLKTGTCKFGATCKFDHPPPG 341
Score = 26.9 bits (58), Expect = 4.1
Identities = 11/20 (55%), Positives = 13/20 (65%), Gaps = 1/20 (5%)
Frame = +3
Query: 271 CENFAK-GTCTFGERCHFAH 289
C+ + K G C FGERC F H
Sbjct: 111 CDFYMKNGECKFGERCKFHH 170
Score = 26.6 bits (57), Expect = 5.4
Identities = 11/23 (47%), Positives = 13/23 (55%), Gaps = 1/23 (4%)
Frame = +3
Query: 270 LCENFAK-GTCTFGERCHFAHGP 291
+C + K GTC FG C F H P
Sbjct: 267 MCPYYLKTGTCKFGATCKFDHPP 335
>TC14620
Length = 1260
Score = 32.7 bits (73), Expect = 0.075
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +3
Query: 264 NNFKTKLCENFAKGTCTFGERCHFAHGPAE 293
+++ C + KG C G+ CHFAHG E
Sbjct: 288 HHYSATPCPDLRKGYCNRGDSCHFAHGVFE 377
Score = 30.0 bits (66), Expect = 0.49
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = +3
Query: 114 CKFGDKCHFAHGEWELGKH 132
C GD CHFAHG +E H
Sbjct: 333 CNRGDSCHFAHGVFECWLH 389
>AV407717
Length = 406
Score = 32.7 bits (73), Expect = 0.075
Identities = 15/32 (46%), Positives = 18/32 (55%), Gaps = 2/32 (6%)
Frame = +1
Query: 266 FKTKLCENFAK--GTCTFGERCHFAHGPAELR 295
+KT LC F G+C+ G C FAH ELR
Sbjct: 205 WKTSLCSYFRSHNGSCSHGTTCRFAHSEEELR 300
Score = 32.0 bits (71), Expect = 0.13
Identities = 15/33 (45%), Positives = 18/33 (54%), Gaps = 1/33 (3%)
Frame = +1
Query: 98 PAVKTRICNKFNTAEG-CKFGDKCHFAHGEWEL 129
P KT +C+ F + G C G C FAH E EL
Sbjct: 199 PLWKTSLCSYFRSHNGSCSHGTTCRFAHSEEEL 297
>TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast nucleoid
DNA binding protein (CND41), partial (43%)
Length = 1002
Score = 32.3 bits (72), Expect = 0.098
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 6/32 (18%)
Frame = +1
Query: 75 APSAQAPPRNV------AAPPPPVPNGSTPAV 100
+PS+ PP+N A+PPPP P+ ++P+V
Sbjct: 847 SPSSNKPPKNTKRNSHTASPPPPAPSATSPSV 942
>AV772854
Length = 494
Score = 32.3 bits (72), Expect = 0.098
Identities = 13/39 (33%), Positives = 19/39 (48%)
Frame = -1
Query: 98 PAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPS 136
P KT +C + C +G +CHF H E K ++ S
Sbjct: 467 PRYKTEVCRMVLSGVVCPYGHRCHFRHALTEQEKAVSQS 351
Score = 32.0 bits (71), Expect = 0.13
Identities = 13/32 (40%), Positives = 17/32 (52%), Gaps = 1/32 (3%)
Frame = -1
Query: 266 FKTKLCENFAKGT-CTFGERCHFAHGPAELRK 296
+KT++C G C +G RCHF H E K
Sbjct: 461 YKTEVCRMVLSGVVCPYGHRCHFRHALTEQEK 366
Score = 29.3 bits (64), Expect = 0.83
Identities = 12/34 (35%), Positives = 17/34 (49%)
Frame = -1
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVA 69
K++ C S CP+G CHF H + AV+
Sbjct: 458 KTEVCRMVLSGVVCPYGHRCHFRHALTEQEKAVS 357
>TC14617 similar to UP|Q42361 (Q42361) Transcribed sequence 1087 protein
(Fragment), partial (31%)
Length = 637
Score = 32.0 bits (71), Expect = 0.13
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 3/63 (4%)
Frame = +3
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIR---EHESDPNLRNIELEGSFDQIKEAS 229
I V G ++G+GG N I + +GAK+ I ++ S R + + GS I+ A
Sbjct: 72 IGVADDHVGLVVGRGGKNIMDISQISGAKIKISDRGDYISGTTDRKVTITGSQRAIRTAE 251
Query: 230 NMV 232
+M+
Sbjct: 252 SMI 260
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,628,089
Number of Sequences: 28460
Number of extensions: 117659
Number of successful extensions: 1581
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 1291
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1521
length of query: 296
length of database: 4,897,600
effective HSP length: 90
effective length of query: 206
effective length of database: 2,336,200
effective search space: 481257200
effective search space used: 481257200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0221.10


