

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
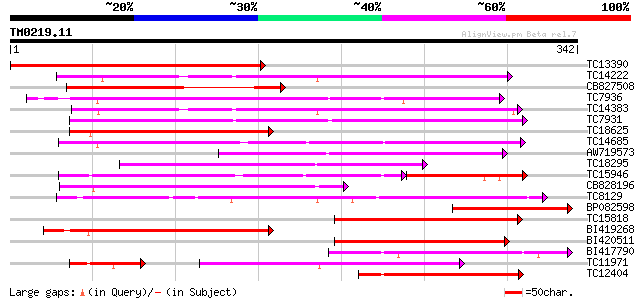
Query= TM0219.11
(342 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13390 weakly similar to UP|O48631 (O48631) Ethylene-forming-en... 306 4e-84
TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carb... 166 5e-42
CB827508 156 5e-39
TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, pa... 152 8e-38
TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-ca... 149 9e-37
TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial... 143 5e-35
TC18625 weakly similar to UP|O48631 (O48631) Ethylene-forming-en... 137 3e-33
TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 132 1e-31
AW719573 128 1e-30
TC18295 similar to UP|Q39224 (Q39224) SRG1 mRNA (F6I1.30/F6I1.30... 128 1e-30
TC15946 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 92 2e-29
CB828196 122 7e-29
TC8129 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (90%) 117 2e-27
BP082598 117 2e-27
TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyge... 117 4e-27
BI419268 111 2e-25
BI420511 108 2e-24
BI417790 102 7e-23
TC11971 weakly similar to UP|Q43419 (Q43419) ACC oxidase, partia... 91 8e-23
TC12404 similar to UP|FLS_CITUN (Q9ZWQ9) Flavonol synthase (FLS... 100 3e-22
>TC13390 weakly similar to UP|O48631 (O48631) Ethylene-forming-enzyme-like
dioxygenase, partial (24%)
Length = 570
Score = 306 bits (783), Expect = 4e-84
Identities = 154/154 (100%), Positives = 154/154 (100%)
Frame = +2
Query: 1 MSKSVQEMSMDSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKLRSALSS 60
MSKSVQEMSMDSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKLRSALSS
Sbjct: 86 MSKSVQEMSMDSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKLRSALSS 265
Query: 61 AGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLD 120
AGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLD
Sbjct: 266 AGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLD 445
Query: 121 WSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
WSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF
Sbjct: 446 WSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 547
>TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carboxylate
oxidase homolog (ACC oxidase) , partial (97%)
Length = 1597
Score = 166 bits (420), Expect = 5e-42
Identities = 89/280 (31%), Positives = 157/280 (55%), Gaps = 5/280 (1%)
Frame = +1
Query: 29 KDSSTLIPIPIIDVSLLSSEDEQGKL---RSALSSAGCFQAIGHGMSSTYLDKIREVAKH 85
++ + P++D+S L++E+ + + A + G F+ + H +S+ ++D + + K
Sbjct: 31 REEKKMANFPVVDMSKLNTEERVATMELIKDACENWGFFELVNHEISTEFMDTVERLTKE 210
Query: 86 FFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPS 145
+ +E++ K A G + V S+ LDW LR P +S P+
Sbjct: 211 HYKRFMEQRFKEMVATK-----GLETVQSEIDDLDWESTFFLRHLPSSN--ISEVPDLDE 369
Query: 146 DFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQF--GEQSSLVARFNFYPPCSRP 203
D+ + + EF+ K++ + ++LL + +L LE+G F + + + + YPPC +P
Sbjct: 370 DYRKVMKEFAVKLEKLAENLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKP 549
Query: 204 DLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGI 263
DL+ G++ HTD GI +L QD +V GLQ+L DD+W++VP + ++VVNLGDQ+++++NG
Sbjct: 550 DLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMRHSIVVNLGDQLEVITNGK 729
Query: 264 FKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINE 303
+KS MHRV+ T+ RMS+A F P + I P L+ E
Sbjct: 730 YKSVMHRVIAQTDGARMSLASFYNPGDDAVISPAPSLVKE 849
>CB827508
Length = 272
Score = 156 bits (394), Expect = 5e-39
Identities = 90/132 (68%), Positives = 90/132 (68%)
Frame = +2
Query: 35 IPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK 94
IPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK
Sbjct: 2 IPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK 181
Query: 95 QKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
QKYARAVNEAE GESLVEF
Sbjct: 182 QKYARAVNEAE------------------------------------------GESLVEF 235
Query: 155 STKVKSMMDHLL 166
STKVKSMMDHLL
Sbjct: 236 STKVKSMMDHLL 271
>TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(94%)
Length = 1492
Score = 152 bits (384), Expect = 8e-38
Identities = 93/295 (31%), Positives = 151/295 (50%), Gaps = 7/295 (2%)
Frame = +3
Query: 11 DSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLSSEDEQ-----GKLRSALSSAGCFQ 65
D DE P V N+F ++ IP+I ++ + D++ K+ A + G FQ
Sbjct: 213 DEDERPK---VAYNNFSNE-------IPVISLAGIDEVDDRRSEICNKIVEACENWGIFQ 362
Query: 66 AIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRL 125
+ HG+ + + + +AK FFALP EEK ++ + G+ + + V DW +
Sbjct: 363 VVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIV 542
Query: 126 SLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG 185
+ +P R S WP+ P+ + E+S K+ + LL ++ ++ LE+ +
Sbjct: 543 TYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACV 722
Query: 186 EQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD--KWVNVPT 243
+ V N+YP C +PDL LG+K HTD IT+LLQD +V GLQ D+ W+ V
Sbjct: 723 DMDQKVV-VNYYPKCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQP 896
Query: 244 IPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVE 298
+ A VVNLGD +SNG FK+ H+ + N+ R+S+A F P P+ + P++
Sbjct: 897 VEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK 1061
>TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1249
Score = 149 bits (375), Expect = 9e-37
Identities = 84/280 (30%), Positives = 153/280 (54%), Gaps = 8/280 (2%)
Frame = +3
Query: 38 PIIDVSLLSSEDEQG---KLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK 94
P+I + L+ E+ + +++ A + G F+ + HG+ +D + + K + + +E++
Sbjct: 51 PVISLEKLNGEERKDIKLQIKDACENWGFFELVNHGIPHDVMDTVERLTKEHYRICMEQR 230
Query: 95 QKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
K A G + V ++ + +DW LR P+ +S P+ ++ + + +F
Sbjct: 231 FKELMASK-----GLEAVQTEVKDMDWESTFHLRHLPESN--ISEIPDLTDEYRKVMKDF 389
Query: 155 STKVKSMMDHLLRTMARSLNLEEGSFLSQF--GEQSSLVARFNFYPPCSRPDLVLGVKPH 212
+ +++ + + LL + +L LE+G F + + YPPC +P+LV G++ H
Sbjct: 390 ALRIEKLSEDLLDLLCENLGLEKGYLKKAFHGSRGPTFGTKVANYPPCPKPNLVKGLRAH 569
Query: 213 TDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVL 272
TD GI +L QD +V GLQ+L D +WV+VP + ++VVNLGDQ+++++NG +KS HRV+
Sbjct: 570 TDAGGIILLFQDDKVSGLQLLKDGEWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVI 749
Query: 273 TNTERLRMSVAMFNEPEPENEIGPVEGLIN---ETRPRLY 309
T+ RMS+A F P + I P L+ E + +LY
Sbjct: 750 AQTDGTRMSIASFYNPGSDAVIYPAPELLEKEAEEKNQLY 869
>TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial (29%)
Length = 1477
Score = 143 bits (360), Expect = 5e-35
Identities = 83/278 (29%), Positives = 142/278 (50%), Gaps = 2/278 (0%)
Frame = +1
Query: 37 IPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEK-Q 95
+P++D+ + ++ A G FQ I HG+S+ ++ + K F A+P EEK
Sbjct: 133 VPVVDLGGHDRAETLKQIFKASEEYGFFQVINHGVSNELIEDTLNIFKEFHAMPAEEKLS 312
Query: 96 KYARAVNEAEGYGNDRVVSKKQVLD-WSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
+ +R N + R ++ K + W L P E+ + WP P+ + E + ++
Sbjct: 313 ESSRDPNGSCMLYTSREINNKDCIQFWKDTLRHPCTPSEEF-MEFWPLKPARYREVVEKY 489
Query: 155 STKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTD 214
+ +++ + +L + L L+ + E L++ N YPPC P L LG+ H D
Sbjct: 490 TKELRELGSQILEMLCEGLGLDPRYCCAGLSENPLLLS--NHYPPCPEPSLTLGLSKHRD 663
Query: 215 RSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTN 274
+ +T+LLQD +V LQV D +W+++ IP A VVN+G +QI+SNG HRV+TN
Sbjct: 664 PTLVTILLQDTDVNALQVFKDGEWISIEPIPYAFVVNIGLLLQIISNGRLVGVEHRVVTN 843
Query: 275 TERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNV 312
+ R +VA F P+ E + P + L+ +YR++
Sbjct: 844 SSISRTTVAYFIRPKKEEIVEPAKALVRTGAHPIYRSI 957
>TC18625 weakly similar to UP|O48631 (O48631) Ethylene-forming-enzyme-like
dioxygenase, partial (33%)
Length = 599
Score = 137 bits (344), Expect = 3e-33
Identities = 64/127 (50%), Positives = 93/127 (72%), Gaps = 4/127 (3%)
Frame = +3
Query: 37 IPIIDVSLLSS----EDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVE 92
IP++D+ LL+S + E KL ALS+ GCFQAI HGM S++LDK+REV+K FF LP E
Sbjct: 216 IPVVDLHLLTSPSTAQQELAKLHYALSTWGCFQAINHGMPSSFLDKVREVSKQFFDLPKE 395
Query: 93 EKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLV 152
EKQKYAR N EGYGND+++ + Q LDW+ R+ L+V P+++R L +WP+ P++F ++
Sbjct: 396 EKQKYAREPNGLEGYGNDQILIQNQRLDWTDRVYLKVQPEDQRNLKVWPQKPNEFRSTIF 575
Query: 153 EFSTKVK 159
E++ +K
Sbjct: 576 EYTKNLK 596
>TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (38%)
Length = 1323
Score = 132 bits (331), Expect = 1e-31
Identities = 92/287 (32%), Positives = 147/287 (51%), Gaps = 5/287 (1%)
Frame = +1
Query: 30 DSSTLIPIPIIDVSLLSSEDEQ--GKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFF 87
+S + IP ID++ ++ ++R A SS G FQ + HG+ L + K F
Sbjct: 277 ESDSKPEIPTIDLAAVNESRAAVVDQIRRAASSVGFFQVVNHGVPLDLLRRTVAAIKAFH 456
Query: 88 ALPVEEKQK-YARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSD 146
P EE+ + Y R + Y ++ + + + W L LR+ P + + P
Sbjct: 457 EQPEEERARVYTREMGTGVSYISNVDLFQSKAASWRDTLQLRMGPTAVDQSMI----PEV 624
Query: 147 FGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLV 206
+ ++E+ +V + + L ++ L L+ G LS+ G V ++YP C +PDL
Sbjct: 625 CRQEVMEWDKEVVRVGEVLYGLLSEGLGLDAGR-LSEMGLVQGRVMVGHYYPFCPQPDLT 801
Query: 207 LGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKS 266
+G+ H D +TV+LQD + GLQV + WV+V + ALV+N+GD +QI+SN +KS
Sbjct: 802 VGLNSHADPGALTVVLQDH-IGGLQVRTTEGWVDVKPLDGALVINIGDLLQIISNEEYKS 978
Query: 267 PMHRVLTNT-ERLRMSVAMFNEP-EPENEIGPVEGLINETRPRLYRN 311
HRVL N+ R+SVA+F P E GP+ L + +P LYRN
Sbjct: 979 ADHRVLANSANEPRVSVAVFLNPGNREKLFGPLPELTSAEKPALYRN 1119
>AW719573
Length = 521
Score = 128 bits (322), Expect = 1e-30
Identities = 61/174 (35%), Positives = 102/174 (58%)
Frame = +2
Query: 127 LRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGE 186
LR P R+ L+ P F E+L + ++ ++ ++ MA +L ++ + E
Sbjct: 2 LRTLPPHIRKPHLFNHIPQPFRENLEIYCAELNNLSIKIMELMANALKIDPKEITEIYNE 181
Query: 187 QSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPD 246
S + R N+YPPC +P+ V+G+K HTD +G+T+LLQ ++EGLQ+ D WV V +P+
Sbjct: 182 GSQTI-RMNYYPPCPQPERVIGLKSHTDGAGLTILLQANDIEGLQIRKDGHWVPVQPLPN 358
Query: 247 ALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGL 300
A ++N+GD ++I +NGI+ S HR N+E+ R+S+A F P + + P L
Sbjct: 359 AFIINIGDMLEITTNGIYASIEHRATVNSEKERISLATFYGPNMQATLAPAPSL 520
>TC18295 similar to UP|Q39224 (Q39224) SRG1 mRNA (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (29%)
Length = 561
Score = 128 bits (322), Expect = 1e-30
Identities = 62/186 (33%), Positives = 109/186 (58%)
Frame = +3
Query: 67 IGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLS 126
I HG++ + ++ +++ + F LP+EEK+K + +G+G VVS++Q L+W+
Sbjct: 6 INHGVNPSLVENMKKGVQDLFNLPIEEKKKLWQTPEVGQGFGQAFVVSEEQKLEWADLFY 185
Query: 127 LRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGE 186
+ FP KR SL P P F ++L + +++K++ ++ M ++L E + F E
Sbjct: 186 INTFPLNKRNTSLIPSIPQPFRDNLDAYCSELKNICVTIIGLMEKALKTETNELVELF-E 362
Query: 187 QSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPD 246
+S R N+YPPC +P+ V+G+ PH+D T+LLQ E+EGLQ+ D WV + + +
Sbjct: 363 DTSQGMRMNYYPPCPQPEKVIGLNPHSDSGAFTILLQVNEMEGLQIRKDGIWVPIKPLSN 542
Query: 247 ALVVNL 252
A V+N+
Sbjct: 543 AFVINI 560
>TC15946 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (56%)
Length = 1252
Score = 91.7 bits (226), Expect(2) = 2e-29
Identities = 64/211 (30%), Positives = 108/211 (50%), Gaps = 1/211 (0%)
Frame = +1
Query: 30 DSSTLIPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFAL 89
++S L +P+ID++ SE GK+RSA G FQ I HG+ ++ LD++ + + F
Sbjct: 208 ENSKLSSVPLIDLTDEHSE-VIGKIRSACHEWGFFQVINHGIPTSVLDEMIDGIRRFHEQ 384
Query: 90 PVE-EKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFG 148
E KQ ++R + + Y + + Q +W V P + E P
Sbjct: 385 DSEVRKQFHSRDLKKKVMYYTNISLFSGQAANWRDTFGFAVAPDPPKP----DEIPLVCR 552
Query: 149 ESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLG 208
+ ++E+S K+K + +L ++ L L S+L + L ++YPPC P L +G
Sbjct: 553 DIVMEYSKKIKELGFTILELLSEGLGLNP-SYLKELNCAEGLFVLGHYYPPCPEPKLTMG 729
Query: 209 VKPHTDRSGITVLLQDREVEGLQVLVDDKWV 239
HTD + IT+LLQD ++ GLQ+L +++WV
Sbjct: 730 TTKHTDGNFITLLLQD-QLGGLQILHENQWV 819
Score = 54.3 bits (129), Expect(2) = 2e-29
Identities = 33/79 (41%), Positives = 49/79 (61%), Gaps = 6/79 (7%)
Frame = +2
Query: 240 NVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMF--NEPEP-ENEI-- 294
+V + ALVVN+GD +Q+++N F S HRVL+ R+SVA F N P+P E +
Sbjct: 821 DVHPVHGALVVNIGDLLQLITNDRFVSVYHRVLSQNIGPRISVASFFVNSPDPIEGTLKV 1000
Query: 295 -GPVEGLINETRPRLYRNV 312
GP++ L+ E P +YR+V
Sbjct: 1001YGPIKELLTEENPPIYRDV 1057
>CB828196
Length = 555
Score = 122 bits (307), Expect = 7e-29
Identities = 66/178 (37%), Positives = 105/178 (58%), Gaps = 4/178 (2%)
Frame = +3
Query: 31 SSTLIPIPIIDVSLLSSED----EQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHF 86
S+T +P+PIID+S L S+D E +L A G FQ I HG++++ L+ ++ + F
Sbjct: 27 STTPLPLPIIDLSKLLSKDHKVPELERLHQACKEWGFFQLINHGVNTSLLEDVKRGVQEF 206
Query: 87 FALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSD 146
F LP+EEK+K+ + +AEGYG VVS+ Q L+W + +FP EKR+ L+P+ PS
Sbjct: 207 FHLPMEEKKKFEQRQGDAEGYGQVFVVSEDQKLEWGDVFFMFLFPLEKRKPYLFPKLPSK 386
Query: 147 FGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPD 204
F + L + T+VK + H+L MA +L ++ ++ R N+YPPC +P+
Sbjct: 387 FRDDLDTYYTEVKKLGIHILELMANALKIDTKEITELL--DTTQATRLNYYPPCPQPE 554
>TC8129 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (90%)
Length = 1278
Score = 117 bits (294), Expect = 2e-27
Identities = 83/308 (26%), Positives = 148/308 (47%), Gaps = 12/308 (3%)
Frame = +2
Query: 29 KDSSTLIPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFA 88
K ++ +P +D LS + + + +A G F+ I H + + + A FF
Sbjct: 173 KPTNLFTEVPEVD---LSHPEAKTLIVNACKEFGFFKIINHDIPMELISNLENEALSFFM 343
Query: 89 LPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPK--EKRRLSLWPENPSD 146
P EK+K ++++ GYG+ R+ K + W L P L L +N +
Sbjct: 344 QPQSEKEK--ASLDDPFGYGSKRI-GKNGDVGWVEYLLFNTNPDVISPNPLLLLEQNQNV 514
Query: 147 FGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQF--GEQSSLVARFNFYPPCSRPD 204
F ++ +++ VK++ +L MA L +E+ + S+ E+S + R N YP C +
Sbjct: 515 FSSAVNDYTLAVKNLSCEILELMADGLGIEQRNVFSRLIRDEKSDCIFRLNHYPACGELE 694
Query: 205 L-------VLGVKPHTDRSGITVLLQDREVEGLQV-LVDDKWVNVPTIPDALVVNLGDQM 256
L ++G HTD I+VL + GLQ+ L D WV++P + +N+GD +
Sbjct: 695 LQAIGGRNLIGFGEHTDPQMISVL-RSNNTSGLQICLRDGTWVSIPPDQTSFFINVGDSL 871
Query: 257 QIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYG 316
Q+M+NG F+S HRVL++T R+S+ F ++ P+ L+++ LY+ +G
Sbjct: 872 QVMTNGSFRSVKHRVLSDTAMSRVSMIYFGGAPLTEKLAPLPSLVSKEEDSLYKEF-TWG 1048
Query: 317 DINYRCYQ 324
+ Y+
Sbjct: 1049EYKKAAYK 1072
>BP082598
Length = 377
Score = 117 bits (294), Expect = 2e-27
Identities = 53/72 (73%), Positives = 63/72 (86%)
Frame = -3
Query: 268 MHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGDINYRCYQEGK 327
MHRV+TNTE+LRMS+ +FN P+ ENEIGPVEGLINETRPRLYRN+ +Y INY CYQEGK
Sbjct: 375 MHRVVTNTEKLRMSLVVFNVPDAENEIGPVEGLINETRPRLYRNIKDYLMINYSCYQEGK 196
Query: 328 IALETVQIAHNS 339
I LET+++ HNS
Sbjct: 195 IPLETIKVTHNS 160
>TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (37%)
Length = 618
Score = 117 bits (292), Expect = 4e-27
Identities = 52/113 (46%), Positives = 78/113 (69%)
Frame = +2
Query: 197 YPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQM 256
YP C +PDL LG+ PH+D G+T+LL D V GLQV + W+ V +P+A ++N+GDQ+
Sbjct: 2 YPKCPQPDLTLGLSPHSDPGGLTILLPDDFVSGLQVRKGNYWITVKPVPNAFIINIGDQI 181
Query: 257 QIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLY 309
Q++SN I+KS HRV+ N+ + R+S+AMF P+ + I P + L+ E +P LY
Sbjct: 182 QVLSNTIYKSVEHRVIVNSNQDRVSLAMFYNPKSDLLIEPAKELVTEEKPALY 340
>BI419268
Length = 536
Score = 111 bits (278), Expect = 2e-25
Identities = 58/141 (41%), Positives = 89/141 (62%), Gaps = 2/141 (1%)
Frame = +1
Query: 21 VERNSFGSKDSSTLIPIPIIDVSLLS--SEDEQGKLRSALSSAGCFQAIGHGMSSTYLDK 78
+E+ ++ S SS IPIID SLLS S++E KL +AL G FQ + HG+ + + +
Sbjct: 121 MEKENYKSHLSSE---IPIIDFSLLSDGSKEELLKLDTALKEWGFFQVVNHGIKTELMQR 291
Query: 79 IREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLS 138
++E+ FF LPVEEK KYA ++ +GYG+ VVS +Q+LDW +L V+P + R+L
Sbjct: 292 MKELTAEFFGLPVEEKNKYAMPPDDIQGYGHTSVVSDEQILDWCDQLIFLVYPTKFRKLQ 471
Query: 139 LWPENPSDFGESLVEFSTKVK 159
WPE P F + + +S+++K
Sbjct: 472 FWPETPEGFKDMIEAYSSEIK 534
>BI420511
Length = 384
Score = 108 bits (269), Expect = 2e-24
Identities = 49/105 (46%), Positives = 73/105 (68%)
Frame = +3
Query: 197 YPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQM 256
YP C +P+LV G++ HTD GI +LLQD +V GLQ+L D +WV+VP + ++V+NLGDQ+
Sbjct: 36 YPACPKPELVKGLRAHTDAGGIILLLQDDKVSGLQLLKDGQWVDVPPMRHSIVINLGDQL 215
Query: 257 QIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLI 301
++++NG +KS HRV+ T+ RMS+A F P + I P L+
Sbjct: 216 EVITNGKYKSVEHRVIAKTDGTRMSIASFYNPANDAVIYPAPALL 350
>BI417790
Length = 610
Score = 102 bits (255), Expect = 7e-23
Identities = 59/151 (39%), Positives = 83/151 (54%), Gaps = 4/151 (2%)
Frame = +3
Query: 193 RFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL--VDDKWVNVPTIPDALVV 250
R N+YPPC PD+VLG H D +T+L QD EV GLQV D +WV + +PDA ++
Sbjct: 9 RLNYYPPCPNPDIVLGCGRHKDSGALTILAQD-EVSGLQVRRKCDGQWVRLNPLPDAYII 185
Query: 251 NLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYR 310
N+GD +Q+ N ++S HRV N E+ R+S F P + P+E L NE P Y
Sbjct: 186 NVGDVIQVWCNDAYESVEHRVKLNPEKARLSYPFFLYPSHYTNVEPLEELTNEENPPKY- 362
Query: 311 NVNNYGD--INYRCYQEGKIALETVQIAHNS 339
N+G + + K+ E VQ+A+ S
Sbjct: 363 TPYNWGKFFVTRKRSNFMKLDEENVQVANTS 455
>TC11971 weakly similar to UP|Q43419 (Q43419) ACC oxidase, partial (69%)
Length = 730
Score = 90.9 bits (224), Expect(2) = 8e-23
Identities = 47/163 (28%), Positives = 91/163 (54%), Gaps = 3/163 (1%)
Frame = +3
Query: 115 KKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLN 174
K++ D + + ++ + K + + +++ ++ K+ + + L M+ +L
Sbjct: 228 KQKASDIDWESAFFIWHRPKSNIKQISNLSEELRQTMDQYIEKLIQLAEDLSGLMSENLG 407
Query: 175 LEEGSFLSQFG--EQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQV 232
LE+ F + ++ + YP C P+LV G++ HTD GI +LLQD +V GL+
Sbjct: 408 LEKDYIKKAFSGSDGPAMGTKVAKYPQCPNPELVRGLREHTDAGGIILLLQDDQVPGLEF 587
Query: 233 LVDDKWVNV-PTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTN 274
D KWV + P+ +A+ +N GDQ++++SNG++KS +HRV+ +
Sbjct: 588 SKDGKWVEIPPSKNNAIFINTGDQIEVLSNGLYKSVVHRVMAD 716
Score = 32.3 bits (72), Expect(2) = 8e-23
Identities = 17/50 (34%), Positives = 30/50 (60%), Gaps = 4/50 (8%)
Frame = +2
Query: 37 IPIIDVSLLSSEDEQGKLRSALSSA----GCFQAIGHGMSSTYLDKIREV 82
IPIID S L+ D++G+ + L A GCF HG+ + ++K++++
Sbjct: 8 IPIIDFSTLNG-DKRGETMALLHEACKNWGCFLIENHGIDKSLMEKVKQL 154
>TC12404 similar to UP|FLS_CITUN (Q9ZWQ9) Flavonol synthase (FLS) (CitFLS)
, partial (31%)
Length = 526
Score = 100 bits (250), Expect = 3e-22
Identities = 49/100 (49%), Positives = 69/100 (69%)
Frame = +2
Query: 211 PHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHR 270
PHTD S +T+LL + EV GLQVL D++W++ IP AL+V++GDQ+QI SNG +KS +HR
Sbjct: 2 PHTDLSSLTILLPN-EVPGLQVLKDERWIDAKYIPHALIVHIGDQIQIASNGRYKSVLHR 178
Query: 271 VLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYR 310
+ ER RMS +F EP E E+GP+ L+ + P Y+
Sbjct: 179 TTVDKERTRMSWPVFLEPPAECEVGPLPQLVTQDNPPKYK 298
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,372,100
Number of Sequences: 28460
Number of extensions: 66973
Number of successful extensions: 401
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 376
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 377
length of query: 342
length of database: 4,897,600
effective HSP length: 91
effective length of query: 251
effective length of database: 2,307,740
effective search space: 579242740
effective search space used: 579242740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0219.11


