

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.28
(278 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
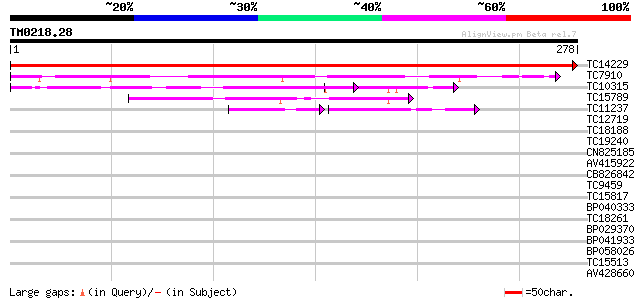
Score E
Sequences producing significant alignments: (bits) Value
TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%) 574 e-165
TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%) 183 2e-47
TC10315 similar to UP|P93166 (P93166) SCOF-1, partial (42%) 137 3e-33
TC15789 similar to UP|O22086 (O22086) ZPT2-14, partial (37%) 70 5e-13
TC11237 similar to UP|Q43614 (Q43614) DNA-binding protein, parti... 59 7e-10
TC12719 homologue to UP|ZFP1_ARATH (Q42485) Zinc finger protein ... 35 0.011
TC18188 similar to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6,... 35 0.018
TC19240 similar to UP|Q94BY4 (Q94BY4) AT5g35570/K2K18_1, partial... 33 0.041
CN825185 33 0.053
AV415922 33 0.070
CB826842 32 0.091
TC9459 similar to AAQ23982 (AAQ23982) Transcription factor Rap21... 32 0.16
TC15817 31 0.20
BP040333 30 0.35
TC18261 similar to UP|WSC2_YEAST (P53832) Cell wall integrity an... 30 0.45
BP029370 30 0.45
BP041933 30 0.45
BP058026 30 0.45
TC15513 homologue to UP|O61085 (O61085) Coronin binding protein,... 30 0.45
AV428660 30 0.45
>TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%)
Length = 1175
Score = 574 bits (1479), Expect = e-165
Identities = 278/278 (100%), Positives = 278/278 (100%)
Frame = +1
Query: 1 MALETLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALC 60
MALETLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALC
Sbjct: 55 MALETLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALC 234
Query: 61 LIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFPSY 120
LIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFPSY
Sbjct: 235 LIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFPSY 414
Query: 121 QALGGHKASHRKSSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKSFPTGQALGGH 180
QALGGHKASHRKSSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKSFPTGQALGGH
Sbjct: 415 QALGGHKASHRKSSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKSFPTGQALGGH 594
Query: 181 KRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHSLRGFDLNLPAPLTEFWTPLGI 240
KRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHSLRGFDLNLPAPLTEFWTPLGI
Sbjct: 595 KRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHSLRGFDLNLPAPLTEFWTPLGI 774
Query: 241 GGGDSRTNKIRSGEQEVESPLPVTAKRPRLFFDGGETA 278
GGGDSRTNKIRSGEQEVESPLPVTAKRPRLFFDGGETA
Sbjct: 775 GGGDSRTNKIRSGEQEVESPLPVTAKRPRLFFDGGETA 888
>TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%)
Length = 1035
Score = 183 bits (465), Expect = 2e-47
Identities = 123/281 (43%), Positives = 152/281 (53%), Gaps = 11/281 (3%)
Frame = +3
Query: 1 MALETLNSPTAAT---TPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDN----PTTTE 53
MA+E L SPT A TPF +E PW K+KRSKR R D+ + TE
Sbjct: 93 MAMEALKSPTTAAPTFTPF------QEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTE 254
Query: 54 EEYLALCLIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVC 113
EEYLALCLIMLA+ + + + S P KL++KCSVC
Sbjct: 255 EEYLALCLIMLARGST------------------AVTPKLTLSRPAPVTAEKLSYKCSVC 380
Query: 114 DKAFPSYQALGGHKASHRK---SSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKS 170
+K FPSYQALGGHKASHRK +++E+ + ++A +AS GGK+H+CSIC KS
Sbjct: 381 EKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSAS-----NGGGKVHQCSICQKS 545
Query: 171 FPTGQALGGHKRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHS-LRGFDLNLPA 229
FPTGQALGGHKRCHYEGG ++ST T T+ G S HS R FDLNLPA
Sbjct: 546 FPTGQALGGHKRCHYEGGGGASST-----------ATATASEGVGSTHSHQRNFDLNLPA 692
Query: 230 PLTEFWTPLGIGGGDSRTNKIRSGEQEVESPLPVTAKRPRL 270
D +K E+EV SPLP +K+PRL
Sbjct: 693 ------------FPDFSASKF-FVEEEVSSPLP--SKKPRL 770
>TC10315 similar to UP|P93166 (P93166) SCOF-1, partial (42%)
Length = 592
Score = 137 bits (344), Expect = 3e-33
Identities = 84/175 (48%), Positives = 103/175 (58%), Gaps = 4/175 (2%)
Frame = +2
Query: 1 MALETLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALC 60
MA+E LNSPT T P S+ + H EPW K+KRSKR R + +EEEYLALC
Sbjct: 140 MAMEALNSPTT-TAP---SFTFNDHPTLRHPAEPWAKRKRSKRSR----SCSEEEYLALC 295
Query: 61 LIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFPSY 120
LIMLA+ G T T L+ ++ + S +L++KCSVCDKAFPSY
Sbjct: 296 LIMLARGGAA------TTTATPPLQPATAPSGSS----------RLSYKCSVCDKAFPSY 427
Query: 121 QALGGHKASHRKSSSENQTPAAAAVNDNASVST----STGNGGKMHECSICHKSF 171
QALGGHKASHRK S +A +A +T +TG+GGK HECSICHKSF
Sbjct: 428 QALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSF 592
Score = 49.7 bits (117), Expect = 6e-07
Identities = 30/72 (41%), Positives = 42/72 (57%), Gaps = 6/72 (8%)
Frame = +2
Query: 155 TGNGGKMHECSICHKSFPTGQALGGHKRCH--YEGG----KSSNSTNSNVHANTSSAVTT 208
+G+ ++CS+C K+FP+ QALGGHK H + GG S+ +T+S V TSSA
Sbjct: 368 SGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSA--N 541
Query: 209 TSDGGAASAHSL 220
T GG A S+
Sbjct: 542 TGSGGKAHECSI 577
>TC15789 similar to UP|O22086 (O22086) ZPT2-14, partial (37%)
Length = 747
Score = 69.7 bits (169), Expect = 5e-13
Identities = 48/146 (32%), Positives = 74/146 (49%), Gaps = 6/146 (4%)
Frame = +1
Query: 59 LCLIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFP 118
L + +LA +N+ + + TN L S Q ++ + + +C C++ F
Sbjct: 109 LFISLLAMKRQRDNEGTESFDLTNCLMLMLSYPKQKRTINES-----VEFECKTCNRKFS 273
Query: 119 SYQALGGHKASHR----KSSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKSFPTG 174
S+QALGGH+ASH K + +TP+ + DN N +MH CS+C F G
Sbjct: 274 SFQALGGHRASHNHKRVKLEEQAKTPS---LWDN--------NKPRMHVCSVCGLGFSLG 420
Query: 175 QALGGHKRCH--YEGGKSSNSTNSNV 198
QALGGH R H EG SS+S++ ++
Sbjct: 421 QALGGHMRKHRNNEGFSSSSSSSYSI 498
>TC11237 similar to UP|Q43614 (Q43614) DNA-binding protein, partial (20%)
Length = 523
Score = 59.3 bits (142), Expect = 7e-10
Identities = 34/74 (45%), Positives = 41/74 (54%)
Frame = +3
Query: 157 NGGKMHECSICHKSFPTGQALGGHKRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAAS 216
N K+HECSIC F +GQALGGH R H ++ TNSN A TS+ V A
Sbjct: 39 NKAKIHECSICGSEFTSGQALGGHMRRHRASANAAAETNSN--AITSTTVV------ARP 194
Query: 217 AHSLRGFDLNLPAP 230
++ DLNLPAP
Sbjct: 195 PRNILQLDLNLPAP 236
Score = 45.1 bits (105), Expect = 1e-05
Identities = 23/47 (48%), Positives = 28/47 (58%)
Frame = +3
Query: 108 HKCSVCDKAFPSYQALGGHKASHRKSSSENQTPAAAAVNDNASVSTS 154
H+CS+C F S QALGGH HR S++ AAA N NA ST+
Sbjct: 54 HECSICGSEFTSGQALGGHMRRHRASAN-----AAAETNSNAITSTT 179
>TC12719 homologue to UP|ZFP1_ARATH (Q42485) Zinc finger protein 1, partial
(18%)
Length = 548
Score = 35.4 bits (80), Expect = 0.011
Identities = 18/53 (33%), Positives = 28/53 (51%)
Frame = +2
Query: 160 KMHECSICHKSFPTGQALGGHKRCHYEGGKSSNSTNSNVHANTSSAVTTTSDG 212
++ C+ CH+ F + QAL G +C EG NS +++SS V T+ G
Sbjct: 395 RVFSCNYCHRKFYSSQALWGSPKCTQEG-----EVNSKKKSSSSSQVWNTNIG 538
>TC18188 similar to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6, partial
(21%)
Length = 594
Score = 34.7 bits (78), Expect = 0.018
Identities = 18/58 (31%), Positives = 29/58 (49%)
Frame = +3
Query: 127 KASHRKSSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKSFPTGQALGGHKRCH 184
KA+ SS++ AA ++ +T +G + +C C + F QALGGH+ H
Sbjct: 216 KATSPSGSSDSGNGDAAG----SASGINTPSGDRKKKCQYCCREFANSQALGGHQNAH 377
Score = 32.7 bits (73), Expect = 0.070
Identities = 12/28 (42%), Positives = 18/28 (63%)
Frame = +3
Query: 109 KCSVCDKAFPSYQALGGHKASHRKSSSE 136
KC C + F + QALGGH+ +H+K +
Sbjct: 312 KCQYCCREFANSQALGGHQNAHKKERQQ 395
>TC19240 similar to UP|Q94BY4 (Q94BY4) AT5g35570/K2K18_1, partial (12%)
Length = 554
Score = 33.5 bits (75), Expect = 0.041
Identities = 23/94 (24%), Positives = 35/94 (36%), Gaps = 3/94 (3%)
Frame = +2
Query: 18 SSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALCLIMLAQSGNNNNKNNNT 77
SS + HH+ +R PRF P T + NNN N+
Sbjct: 77 SSLMASFSDSNHHHNTSDGVAQRVNSPRFSGPMTRRAHSFK------RNNNNNNGNNSGN 238
Query: 78 DNNTNTLRAESSSNSQSQSPPQQ---QPPLKLTH 108
+N+N+L + Q SP + +P L+ H
Sbjct: 239 SSNSNSLSTHNEVELQINSPRSEEGLEPVLERKH 340
>CN825185
Length = 541
Score = 33.1 bits (74), Expect = 0.053
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +3
Query: 110 CSVCDKAFPSYQALGGHKASHRKSSSENQ 138
C C K F S QA+GGH+ +HR E++
Sbjct: 204 CLYCSKRFSSVQAVGGHQNAHRTERVESE 290
Score = 28.5 bits (62), Expect = 1.3
Identities = 11/27 (40%), Positives = 15/27 (54%)
Frame = +3
Query: 158 GGKMHECSICHKSFPTGQALGGHKRCH 184
G + C C K F + QA+GGH+ H
Sbjct: 186 GSHGYGCLYCSKRFSSVQAVGGHQNAH 266
>AV415922
Length = 422
Score = 32.7 bits (73), Expect = 0.070
Identities = 28/93 (30%), Positives = 42/93 (45%)
Frame = +1
Query: 5 TLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALCLIML 64
TL SP++ + F SS + + LH + SKR D+ ++ ++ + +
Sbjct: 28 TLPSPSSPFSIFPSS-RLSPSPKSLHF------RLASKR---DDGQDSDTQFNSKGINTN 177
Query: 65 AQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSP 97
NNNN NNN +NN N L S S SP
Sbjct: 178 NDDNNNNNNNNNNNNNNNNLPILSDRRCFSLSP 276
>CB826842
Length = 637
Score = 32.3 bits (72), Expect = 0.091
Identities = 21/62 (33%), Positives = 32/62 (50%), Gaps = 10/62 (16%)
Frame = +1
Query: 86 AESSSNSQS---QSPPQQQPPLKLTHKCSVCDKAFPSYQALGGH-------KASHRKSSS 135
A++SS SQ +S P ++ C+ C + F + QALGGH +A ++SS
Sbjct: 52 AQNSSKSQQIGWRSDDLGGPGQVRSYSCTFCKRGFSNAQALGGHMNIHTRDRAKLKQSSE 231
Query: 136 EN 137
EN
Sbjct: 232 EN 237
Score = 30.8 bits (68), Expect = 0.27
Identities = 13/40 (32%), Positives = 20/40 (49%), Gaps = 1/40 (2%)
Frame = +1
Query: 160 KMHECSICHKSFPTGQALGGHKRCH-YEGGKSSNSTNSNV 198
+ + C+ C + F QALGGH H + K S+ N+
Sbjct: 121 RSYSCTFCKRGFSNAQALGGHMNIHTRDRAKLKQSSEENL 240
>TC9459 similar to AAQ23982 (AAQ23982) Transcription factor Rap212, partial
(12%)
Length = 434
Score = 31.6 bits (70), Expect = 0.16
Identities = 12/38 (31%), Positives = 24/38 (62%)
Frame = +2
Query: 66 QSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPP 103
+S NNNN NNN+++N+N+ + + ++ + +Q P
Sbjct: 257 ESNNNNNNNNNSNSNSNSNKKKQQKRAKKKYRGVRQRP 370
Score = 26.9 bits (58), Expect = 3.8
Identities = 18/51 (35%), Positives = 24/51 (46%)
Frame = +2
Query: 51 TTEEEYLALCLIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQ 101
T +E +A CL +N NNN +NN+N SNS S QQ+
Sbjct: 200 TCKECNIAGCLGCKFFPEEESNNNNNNNNNSN-------SNSNSNKKKQQK 331
Score = 26.6 bits (57), Expect = 5.0
Identities = 9/36 (25%), Positives = 22/36 (61%)
Frame = +2
Query: 65 AQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQ 100
+ + NNNN N+N+++N+N + + + + + Q+
Sbjct: 260 SNNNNNNNNNSNSNSNSNKKKQQKRAKKKYRGVRQR 367
>TC15817
Length = 773
Score = 31.2 bits (69), Expect = 0.20
Identities = 35/151 (23%), Positives = 52/151 (34%), Gaps = 3/151 (1%)
Frame = +3
Query: 117 FPSYQALGGHKASHRKSSSENQTP--AAAAVNDNASVSTSTGNGGKMHEC-SICHKSFPT 173
FPS+Q G S R + +TP ++ A+ +G GKM + S KS +
Sbjct: 303 FPSHQLSNGLYVSGRPEQPKERTPTMSSVAMPYTGGDIKKSGELGKMFDIPSDGSKSRKS 482
Query: 174 GQALGGHKRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHSLRGFDLNLPAPLTE 233
G R G +S+S +A SA T+ + A PL +
Sbjct: 483 GPLSNAPSRTGSFAGAASHSGPIMQNAAARSAYVTSGNVAAGGMSGSASMKKTNSGPLNK 662
Query: 234 FWTPLGIGGGDSRTNKIRSGEQEVESPLPVT 264
P+ G R + LP T
Sbjct: 663 HGEPIKKSSGPQSGGGTRQNSGPIPPVLPTT 755
>BP040333
Length = 480
Score = 30.4 bits (67), Expect = 0.35
Identities = 12/22 (54%), Positives = 15/22 (67%)
Frame = +1
Query: 67 SGNNNNKNNNTDNNTNTLRAES 88
+ NNNN NNN +NN +LR S
Sbjct: 73 TNNNNNNNNNNNNNNKSLREVS 138
Score = 28.9 bits (63), Expect = 1.0
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +1
Query: 59 LCLIMLAQSGNNNNKNNNTDNNTNTLRAESS 89
L + ++ NNNN NNN +NN N E S
Sbjct: 46 LSWFLKTKTTNNNNNNNNNNNNNNKSLREVS 138
Score = 28.1 bits (61), Expect = 1.7
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +1
Query: 56 YLALCLIMLAQSGNNNNKNNNTDNN 80
YL+ L + NNNN NNN +NN
Sbjct: 43 YLSWFLKTKTTNNNNNNNNNNNNNN 117
>TC18261 similar to UP|WSC2_YEAST (P53832) Cell wall integrity and stress
response component 2 precursor, partial (5%)
Length = 538
Score = 30.0 bits (66), Expect = 0.45
Identities = 27/119 (22%), Positives = 47/119 (38%), Gaps = 2/119 (1%)
Frame = +3
Query: 41 SKRPRFDNPTTTEEEYLALCLIMLAQSGNNNNKNNNTDNNT--NTLRAESSSNSQSQSPP 98
S P NP E + + GN+ + N N+ N L A S + PP
Sbjct: 57 SADPLLKNPAARE-----FSSALRSNRGNSRSFANKRSANSKPNPLNAAQSPPTSPPQPP 221
Query: 99 QQQPPLKLTHKCSVCDKAFPSYQALGGHKASHRKSSSENQTPAAAAVNDNASVSTSTGN 157
QPP L+ S + S + ++S +SS + +P++ S +++T +
Sbjct: 222 PLQPPTSLSLMTSTTSTSSSSPNS----RSSAASASSSSPSPSSVKTTVPGSTASNTSS 386
>BP029370
Length = 364
Score = 30.0 bits (66), Expect = 0.45
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = +1
Query: 59 LCLIMLAQSGNNNNKNNNTDNNTNTL 84
L +I + + NNNNKNNN +NN + +
Sbjct: 154 LHII*IYNNNNNNNKNNNNNNNNSII 231
Score = 27.7 bits (60), Expect = 2.2
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = +1
Query: 56 YLALCLIMLAQSGNNNNKNNNTDNNTNTL 84
Y + I+ + NNNN NN +NN N++
Sbjct: 142 YKRILHII*IYNNNNNNNKNNNNNNNNSI 228
>BP041933
Length = 472
Score = 30.0 bits (66), Expect = 0.45
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 20/98 (20%)
Frame = -1
Query: 62 IMLAQSGNNNNKNNNTDNNT----------NTL----------RAESSSNSQSQSPPQQQ 101
++ +Q NNNN NNN+ T NT+ A +++NS+S SP Q
Sbjct: 412 LVKSQKDNNNNNNNNSITETKSRTDPKPF*NTVSS*NP*PAPNEASNTNNSRSNSP--QI 239
Query: 102 PPLKLTHKCSVCDKAFPSYQALGGHKASHRKSSSENQT 139
LKL H + K + A H+ KS + +T
Sbjct: 238 SALKLIHNAAP-SKPLSKFPAT--HQTKRAKSPTRKKT 134
>BP058026
Length = 389
Score = 30.0 bits (66), Expect = 0.45
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = +3
Query: 69 NNNNKNNNTDNNTNTLRAESSSNS 92
NNNN NNN +NN N +++N+
Sbjct: 12 NNNNNNNNNNNNNNNNNNNNNNNN 83
Score = 30.0 bits (66), Expect = 0.45
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = +3
Query: 69 NNNNKNNNTDNNTNTLRAESSSNS 92
NNNN NNN +NN N +++N+
Sbjct: 9 NNNNNNNNNNNNNNNNNNNNNNNN 80
Score = 30.0 bits (66), Expect = 0.45
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = +3
Query: 67 SGNNNNKNNNTDNNTNTLRAESSSN 91
+ NNNN NNN +NN N +++N
Sbjct: 9 NNNNNNNNNNNNNNNNNNNNNNNNN 83
Score = 29.6 bits (65), Expect = 0.59
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +3
Query: 67 SGNNNNKNNNTDNNTNTL 84
+ NNNN NNN +NN N L
Sbjct: 36 NNNNNNNNNNNNNNNNDL 89
Score = 28.9 bits (63), Expect = 1.0
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +3
Query: 63 MLAQSGNNNNKNNNTDNNTN 82
++ + NNNN NNN +NN N
Sbjct: 3 IINNNNNNNNNNNNNNNNNN 62
Score = 28.1 bits (61), Expect = 1.7
Identities = 10/25 (40%), Positives = 16/25 (64%)
Frame = +3
Query: 70 NNNKNNNTDNNTNTLRAESSSNSQS 94
NNN NNN +NN N +++N+ +
Sbjct: 9 NNNNNNNNNNNNNNNNNNNNNNNNN 83
Score = 26.9 bits (58), Expect = 3.8
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = +3
Query: 67 SGNNNNKNNNTDNNTNTL 84
+ NNNN NNN +NN + +
Sbjct: 39 NNNNNNNNNNNNNNNDLI 92
>TC15513 homologue to UP|O61085 (O61085) Coronin binding protein, partial
(5%)
Length = 1029
Score = 30.0 bits (66), Expect = 0.45
Identities = 20/92 (21%), Positives = 40/92 (42%), Gaps = 5/92 (5%)
Frame = +1
Query: 5 TLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALCLIML 64
T P S Y+ ++++++ H P S+ + PTTT + +
Sbjct: 202 TTTVPITNNLHHHSLYRTQQQQQQHHQSSPIFLDPSSQFIQRQTPTTTTTTFTGKRTLAE 381
Query: 65 AQSGNN-----NNKNNNTDNNTNTLRAESSSN 91
Q+ ++ NN N+N+++N+N + SN
Sbjct: 382 FQTQHHQNQIFNNNNSNSNSNSNFTHNQFLSN 477
Score = 28.9 bits (63), Expect = 1.0
Identities = 21/83 (25%), Positives = 33/83 (39%), Gaps = 15/83 (18%)
Frame = +3
Query: 28 ELHHHEPWTKKKRSKRP----RFDNPTTTEE-----EYLALCLIMLAQSGNNNNKNNNTD 78
++HH +P + ++P NP TT +++ I + + N NN NNN
Sbjct: 180 QIHHQQPHHHRSHHQQPPPPLSLPNPATTTTTSSILSHISRSFIPIHSTPNTNNNNNNLH 359
Query: 79 NNT------NTLRAESSSNSQSQ 95
T NT E + Q Q
Sbjct: 360 R*THLG*IPNTTPPEPNLQQQQQ 428
>AV428660
Length = 408
Score = 30.0 bits (66), Expect = 0.45
Identities = 22/99 (22%), Positives = 40/99 (40%), Gaps = 3/99 (3%)
Frame = +1
Query: 14 TPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALCLIMLAQSGNNNNK 73
TP ++Q + H PWT K + +F + T + L + S NNNN
Sbjct: 64 TPRYQAHQWRNPRVDHHPSPPWTLKLKFPT-QFPHLQTLQSNPLLQSQTPTSPSNNNNNP 240
Query: 74 NNNTDNNTNTLRAESSSNSQSQSP---PQQQPPLKLTHK 109
++ ++ + + +S+N + P PP T +
Sbjct: 241 LHSLKSHKTSYSSHNSNNRRKGKP*I*TLMSPPFNSTKR 357
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.307 0.123 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,239,890
Number of Sequences: 28460
Number of extensions: 81978
Number of successful extensions: 1057
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 826
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 936
length of query: 278
length of database: 4,897,600
effective HSP length: 89
effective length of query: 189
effective length of database: 2,364,660
effective search space: 446920740
effective search space used: 446920740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0218.28


