

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
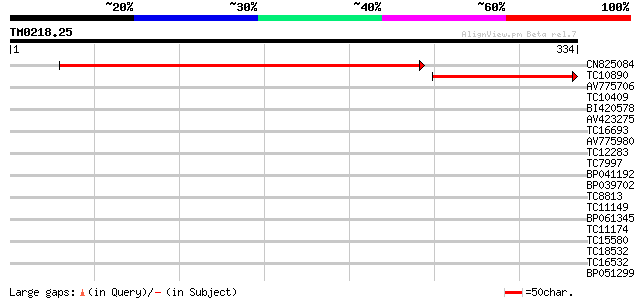
Query= TM0218.25
(334 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CN825084 454 e-129
TC10890 similar to UP|Q93VN2 (Q93VN2) At2g14841/At2g14841, parti... 172 5e-44
AV775706 33 0.067
TC10409 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (17%) 32 0.20
BI420578 30 0.43
AV423275 30 0.74
TC16693 weakly similar to PIR|T51842|T51842 RING-H2 finger prote... 30 0.74
AV775980 29 0.97
TC12283 similar to UP|AAR23700 (AAR23700) At1g65040, partial (21%) 29 0.97
TC7997 similar to UP|Q9ZTM6 (Q9ZTM6) PGPD14, partial (27%) 29 1.3
BP041192 28 1.7
BP039702 28 1.7
TC8813 weakly similar to UP|Q9XF65 (Q9XF65) RING-H2 zinc finger ... 28 2.2
TC11149 28 2.2
BP061345 28 2.2
TC11174 weakly similar to UP|P93837 (P93837) Amp-binding protein... 28 2.2
TC15580 28 2.2
TC18532 similar to UP|AAR00327 (AAR00327) Rapid alkalinization f... 28 2.8
TC16532 similar to UP|Q9LSV4 (Q9LSV4) Gb|AAB61039.1, partial (9%) 28 2.8
BP051299 27 3.7
>CN825084
Length = 664
Score = 454 bits (1168), Expect = e-129
Identities = 215/215 (100%), Positives = 215/215 (100%)
Frame = +3
Query: 30 EHQICVIRTYSEWVIDGEYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKS 89
EHQICVIRTYSEWVIDGEYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKS
Sbjct: 18 EHQICVIRTYSEWVIDGEYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKS 197
Query: 90 FPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVT 149
FPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVT
Sbjct: 198 FPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVT 377
Query: 150 ESRSPPPAFASEPLIGRENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPVG 209
ESRSPPPAFASEPLIGRENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPVG
Sbjct: 378 ESRSPPPAFASEPLIGRENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPVG 557
Query: 210 PATRKGAFNVERQNSEISYYADDEDGNRKKYTKRG 244
PATRKGAFNVERQNSEISYYADDEDGNRKKYTKRG
Sbjct: 558 PATRKGAFNVERQNSEISYYADDEDGNRKKYTKRG 662
>TC10890 similar to UP|Q93VN2 (Q93VN2) At2g14841/At2g14841, partial (24%)
Length = 571
Score = 172 bits (437), Expect = 5e-44
Identities = 85/85 (100%), Positives = 85/85 (100%)
Frame = +2
Query: 250 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACM 309
FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACM
Sbjct: 2 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACM 181
Query: 310 ATMGILYYRLVQRGPGEELPNEEQI 334
ATMGILYYRLVQRGPGEELPNEEQI
Sbjct: 182 ATMGILYYRLVQRGPGEELPNEEQI 256
>AV775706
Length = 462
Score = 33.1 bits (74), Expect = 0.067
Identities = 17/53 (32%), Positives = 24/53 (45%), Gaps = 1/53 (1%)
Frame = -2
Query: 57 CQAVLEEGDGSQTT-RLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
C LE+ Q L C H H+ CL+ +++ PP CP C TS+
Sbjct: 293 CAVCLEDFQPKQQVMNLSCSHTYHSACLLPWLEAHPP--------CPCCRTSV 159
>TC10409 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (17%)
Length = 715
Score = 31.6 bits (70), Expect = 0.20
Identities = 17/56 (30%), Positives = 25/56 (44%)
Frame = +3
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
+CC C + E DG++ L C H H+ C+V +K CP C +I
Sbjct: 42 ECCICISSYE--DGAELHSLPCNHHFHSTCIVKWLK--------MNATCPLCKYNI 179
>BI420578
Length = 397
Score = 30.4 bits (67), Expect = 0.43
Identities = 22/53 (41%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Frame = +1
Query: 255 LPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSS--RMDPRKILLLIAI 305
LPF+S +LP LP +PP E H+RSS R+ PR LL+ AI
Sbjct: 49 LPFFSPSLPWLPRLSPP----------PETTNHHRRSSSQRILPRSPLLIGAI 177
>AV423275
Length = 405
Score = 29.6 bits (65), Expect = 0.74
Identities = 27/95 (28%), Positives = 39/95 (40%), Gaps = 15/95 (15%)
Frame = +2
Query: 155 PPAFASEPLIGRENHGNSDSPA--TGSEPPKLS-----------VTDIVEIEGAN--SAG 199
PP F S P++ R + +S +PA S P +S V + + G +A
Sbjct: 53 PPPFNSPPVLPRAHTCHSLAPARVVQSSPSSISRFEFRVFQQPCVKSLRAVRGGGVVTAV 232
Query: 200 NFVKGSSPVGPATRKGAFNVERQNSEISYYADDED 234
KG VG F+ E + I YY DD+D
Sbjct: 233 AKKKGRGGVGIVGVDEGFDEEEEGDGIEYYDDDDD 337
>TC16693 weakly similar to PIR|T51842|T51842 RING-H2 finger protein RHA2a
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (35%)
Length = 442
Score = 29.6 bits (65), Expect = 0.74
Identities = 16/58 (27%), Positives = 24/58 (40%)
Frame = +2
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWP 110
+C C + EEG+ + RL C H H +CL ++ CP C + P
Sbjct: 230 ECRVCLSEFEEGE--KVRRLKCKHTFHKDCLDKWLQE-------CWATCPLCRKQVLP 376
>AV775980
Length = 480
Score = 29.3 bits (64), Expect = 0.97
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +2
Query: 5 KCRKATKLYCFVHKVPVCGECICFPEHQI 33
+CRK ++ C VCG+CI F E I
Sbjct: 92 RCRKTSQFRCLCCPSAVCGKCIYFGEFAI 178
>TC12283 similar to UP|AAR23700 (AAR23700) At1g65040, partial (21%)
Length = 390
Score = 29.3 bits (64), Expect = 0.97
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = +2
Query: 71 RLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKS 113
+L C H+ H +CL S ++ HT CPTC + PP++
Sbjct: 173 KLICGHLFHVHCLRSWLER--QHT------CPTCRALVVPPEN 277
>TC7997 similar to UP|Q9ZTM6 (Q9ZTM6) PGPD14, partial (27%)
Length = 635
Score = 28.9 bits (63), Expect = 1.3
Identities = 15/42 (35%), Positives = 20/42 (46%)
Frame = +2
Query: 67 SQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
+ T + C H IH +CL + F YACP CS S+
Sbjct: 14 NDVTVMLCGHTIHKSCLNEMREHFQ-------YACPLCSKSV 118
>BP041192
Length = 503
Score = 28.5 bits (62), Expect = 1.7
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 15/92 (16%)
Frame = +3
Query: 108 IWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVS--------------LSVTESRS 153
+WPP + D+ S + L++ M G IF V LS E +S
Sbjct: 39 LWPPVNKADNPSLHNPLLRQQRMGCGSLAAIFELEGVLIEDNLDLENQAWLVLSQQEGKS 218
Query: 154 PPPAFASEPLIG-RENHGNSDSPATGSEPPKL 184
PPPAF + + G + H S+ +P ++
Sbjct: 219 PPPAFILKRIEGMKSEHAVSEVLCWSXDPAQV 314
>BP039702
Length = 553
Score = 28.5 bits (62), Expect = 1.7
Identities = 16/42 (38%), Positives = 22/42 (52%)
Frame = +3
Query: 167 ENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPV 208
+NHG S SP+T + PP+ +I A +A K SS V
Sbjct: 219 KNHGVSASPSTANSPPEKVKEEITTTATATAAACNAKLSSEV 344
>TC8813 weakly similar to UP|Q9XF65 (Q9XF65) RING-H2 zinc finger protein
ATL6, partial (24%)
Length = 495
Score = 28.1 bits (61), Expect = 2.2
Identities = 16/37 (43%), Positives = 20/37 (53%), Gaps = 6/37 (16%)
Frame = +1
Query: 56 QCQAVLEEGDGSQTTRL--GCLHVIHTNC----LVSH 86
+C L E + ++T RL C HV HT C LVSH
Sbjct: 373 ECAVCLSEFEDAETLRLIPKCDHVFHTECIDEWLVSH 483
>TC11149
Length = 881
Score = 28.1 bits (61), Expect = 2.2
Identities = 14/45 (31%), Positives = 18/45 (39%)
Frame = +2
Query: 60 VLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTC 104
++ GD L C HV H CL + P T + CP C
Sbjct: 236 IVRSGDMPTAGILPCRHVFHAECL----EQTTPKTRKSDPPCPVC 358
>BP061345
Length = 543
Score = 28.1 bits (61), Expect = 2.2
Identities = 13/32 (40%), Positives = 16/32 (49%)
Frame = +2
Query: 80 TNCLVSHIKSFPPHTAPAGYACPTCSTSIWPP 111
T+C +HI F P AP A P C + PP
Sbjct: 89 TSCSSNHITFFFPDIAPMVTASPVCPWACTPP 184
>TC11174 weakly similar to UP|P93837 (P93837) Amp-binding protein, partial
(15%)
Length = 689
Score = 28.1 bits (61), Expect = 2.2
Identities = 11/27 (40%), Positives = 17/27 (62%), Gaps = 4/27 (14%)
Frame = -2
Query: 91 PPHTAPA----GYACPTCSTSIWPPKS 113
PP T+P+ AC +C T ++PP+S
Sbjct: 217 PPKTSPSWEP*SVACSSCKTQLYPPQS 137
>TC15580
Length = 719
Score = 28.1 bits (61), Expect = 2.2
Identities = 18/61 (29%), Positives = 27/61 (43%)
Frame = -2
Query: 123 SKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSDSPATGSEPP 182
S KE + + M++++ N P L S SP FAS G + SP +PP
Sbjct: 382 SLTKEQLCRKHMQQHVSANRPQQLEAAFSNSPSDGFASFIAGGMFSVVAFKSPKPRLKPP 203
Query: 183 K 183
+
Sbjct: 202 R 200
>TC18532 similar to UP|AAR00327 (AAR00327) Rapid alkalinization factor 3,
partial (41%)
Length = 580
Score = 27.7 bits (60), Expect = 2.8
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 7/103 (6%)
Frame = -2
Query: 119 SRLH--SKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSDSPA 176
SR H +++ I+ +E N P SLS++ + S PPA + + R + S
Sbjct: 519 SREHELTQILNFILDLRVESNRKKKMPTSLSLSHTHSLPPAASDDRASPRVRVDRASS-- 346
Query: 177 TGSEPPKLSVTDIVEIEGANSAGNFVKGSSPVG-----PATRK 214
++V +V AG+ V G S VG P+ RK
Sbjct: 345 ------LVAVMSVVRSTRTRRAGDPVGGESAVGNVVLDPSARK 235
>TC16532 similar to UP|Q9LSV4 (Q9LSV4) Gb|AAB61039.1, partial (9%)
Length = 471
Score = 27.7 bits (60), Expect = 2.8
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +2
Query: 122 HSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEP 162
HS + + +KN+F N +SLS+ +P P+ A EP
Sbjct: 152 HSATQPLASSSSTKKNLFKNPILSLSLINLFTPFPSLAVEP 274
>BP051299
Length = 539
Score = 27.3 bits (59), Expect = 3.7
Identities = 18/77 (23%), Positives = 30/77 (38%)
Frame = +3
Query: 110 PPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENH 169
PP S + H + T + I P + + S +PPP+ + P
Sbjct: 102 PPPPSSPSTATTHPPPSSSSSATSVLFPISSTTPSAATTLFSPNPPPSASPPPSTVNSPS 281
Query: 170 GNSDSPATGSEPPKLSV 186
S + ++ S PPK +V
Sbjct: 282 SESVTASSASPPPKTAV 332
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,542,334
Number of Sequences: 28460
Number of extensions: 136818
Number of successful extensions: 883
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 868
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 882
length of query: 334
length of database: 4,897,600
effective HSP length: 91
effective length of query: 243
effective length of database: 2,307,740
effective search space: 560780820
effective search space used: 560780820
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0218.25


