

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.19
(794 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
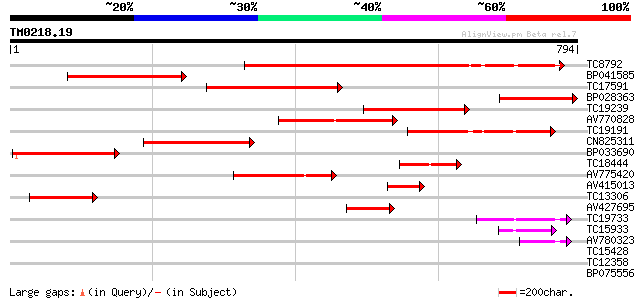
Score E
Sequences producing significant alignments: (bits) Value
TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosid... 384 e-107
BP041585 354 3e-98
TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (... 233 7e-62
BP028363 218 4e-57
TC19239 homologue to UP|Q9ZU04 (Q9ZU04) Beta-glucosidase (Fragm... 186 9e-48
AV770828 180 9e-46
TC19191 similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragme... 176 2e-44
CN825311 174 5e-44
BP033690 158 3e-39
TC18444 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial... 132 2e-31
AV775420 131 5e-31
AV415013 108 3e-24
TC13306 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (12%) 108 3e-24
AV427695 77 1e-14
TC19733 weakly similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11,... 72 3e-13
TC15933 weakly similar to UP|BAC98299 (BAC98299) LEXYL2 protein ... 58 5e-09
AV780323 44 8e-05
TC15428 38 0.005
TC12358 weakly similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, pa... 34 0.10
BP075556 32 0.30
>TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase
{Prunus persica;} , partial (90%)
Length = 1636
Score = 384 bits (985), Expect = e-107
Identities = 210/450 (46%), Positives = 280/450 (61%), Gaps = 2/450 (0%)
Frame = +3
Query: 330 LRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFFDN-PKSLPFANLGPSDVCTKENQL 388
L +T A+K + + ++ AL V MRLG FD P + P+ NLGP DVCT +
Sbjct: 3 LALHTDAAIKNGLITENDLNLALANLITVQMRLGMFDGEPSAHPYGNLGPRDVCTPAHNE 182
Query: 389 LALEAAKQGIVLLENSG-VLPLSKTNIKNLAVIGPNANATTVMISNYAGIPCRYTSPLQG 447
LALEAA+QGIVLLEN G LPLS T + + VIGPN++ T MI NYAG+ C YT+PLQG
Sbjct: 183 LALEAARQGIVLLENKGNALPLSPTRHRTVGVIGPNSDVTVTMIGNYAGVACGYTTPLQG 362
Query: 448 LQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSIEAEGLDRVNLTL 507
+ +Y+ ++ A GC +V C LF A A +DA V+VVGLDQSIEAE DRV L L
Sbjct: 363 IARYVKTIHQA-GCRDVSCRGTELFGNAEIVARQSDATVVVVGLDQSIEAEFRDRVGLLL 539
Query: 508 PGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQV 567
PG Q++LV VA A +G V+LVIM+ GP+D+ F K I ILWVGYPGQAGG AIA V
Sbjct: 540 PGHQQELVSRVARAARGPVVLVIMSGGPVDVQFAKDDPKISAILWVGYPGQAGGTAIADV 719
Query: 568 IFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLS 627
IFG NPGGR P TWYPQSY ++PMT+M+MR N + +PGRTYRFY G ++ FGHGLS
Sbjct: 720 IFGRTNPGGRLPNTWYPQSYVAKLPMTNMDMRPNPASGYPGRTYRFYKGPVVFPFGHGLS 899
Query: 628 YSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSDGQAIDISTISCLDLTFFLVIGVKN 687
YS F+ +A AP + + P S+ N+ S +A+ +S C L + VKN
Sbjct: 900 YSRFTHTLAVAPEQVSV-PIASLQAFRNSTMSS----KAVKVSHAKCGALKMGFHVDVKN 1064
Query: 688 TGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSNVD 747
G +G+H +L+F PP + + KQL+ F + +V G + V + + +C+ LS VD
Sbjct: 1065EGSMDGTHTLLIFSTPPPGK---WSASKQLVSFHKTYVPAGSKQRVNVGVHVCKHLSVVD 1235
Query: 748 SDGKRKLIIGKHTLLVGSSSETQVRHQIDI 777
+G R++ +G H L +G V+H I +
Sbjct: 1236ENGIRRIPMGDHELHIG-----DVKHTISV 1310
>BP041585
Length = 503
Score = 354 bits (908), Expect = 3e-98
Identities = 167/167 (100%), Positives = 167/167 (100%)
Frame = +1
Query: 81 PSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARA 140
PSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARA
Sbjct: 1 PSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARA 180
Query: 141 MYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVEDEESIKA 200
MYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVEDEESIKA
Sbjct: 181 MYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVEDEESIKA 360
Query: 201 DRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKS 247
DRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKS
Sbjct: 361 DRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKS 501
>TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (24%)
Length = 582
Score = 233 bits (595), Expect = 7e-62
Identities = 119/193 (61%), Positives = 147/193 (75%), Gaps = 2/193 (1%)
Frame = +3
Query: 276 LKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTATPEDAVALALKAGLNMNCGDYLRKYTA 335
LKGVIRGQW L+GYIVSDCDSV+V ++ HYT TPE+A A ++ AGL+++CG YL KYT
Sbjct: 3 LKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE 182
Query: 336 NAVKLKKVDVSTVDQALVYNYIVLMRLGFFD-NPKSLPFANLGPSDVCTKENQLLALEAA 394
AVK +D ++++ A+ N+ LMRLGFFD NP +LP+ LGP DVCT ENQ LA EAA
Sbjct: 183 GAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAA 362
Query: 395 KQGIVLLENS-GVLPLSKTNIKNLAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYIS 453
+QGIVLL+NS G LPLS +IK+LAVIGPNANAT VMI N GIPC+Y SPLQGL +
Sbjct: 363 RQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGN*EGIPCKYISPLQGLTALVP 542
Query: 454 SVTYAPGCGNVKC 466
+ +Y PGC +V C
Sbjct: 543 T-SYVPGCPDVHC 578
>BP028363
Length = 508
Score = 218 bits (554), Expect = 4e-57
Identities = 108/109 (99%), Positives = 109/109 (99%)
Frame = -3
Query: 686 KNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSN 745
KNTGP+NGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSN
Sbjct: 506 KNTGPYNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSN 327
Query: 746 VDSDGKRKLIIGKHTLLVGSSSETQVRHQIDISLSGNGMEKKSYLDESI 794
VDSDGKRKLIIGKHTLLVGSSSETQVRHQIDISLSGNGMEKKSYLDESI
Sbjct: 326 VDSDGKRKLIIGKHTLLVGSSSETQVRHQIDISLSGNGMEKKSYLDESI 180
>TC19239 homologue to UP|Q9ZU04 (Q9ZU04) Beta-glucosidase (Fragment) ,
partial (69%)
Length = 453
Score = 186 bits (473), Expect = 9e-48
Identities = 86/149 (57%), Positives = 109/149 (72%)
Frame = +1
Query: 496 EAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGY 555
+AE LDRVN+ LPG Q+ LV +VA+A++G VILVIM+ G +D+SF K+ I ILWVGY
Sbjct: 1 QAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGY 180
Query: 556 PGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYN 615
PG+AGG AIA VIFG YNP GR P TWYP+SY + VPMT+MNMRA+ S +PGRTYRFY
Sbjct: 181 PGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYK 360
Query: 616 GNSLYEFGHGLSYSTFSTYIASAPSTIMI 644
G +++ FG G+ YS I AP + +
Sbjct: 361 GETVFSFGDGIGYSNVEHKIVKAPQLVFV 447
>AV770828
Length = 527
Score = 180 bits (456), Expect = 9e-46
Identities = 98/168 (58%), Positives = 120/168 (71%), Gaps = 1/168 (0%)
Frame = +1
Query: 377 GPSDVCTKENQLLALEAAKQGIVLLENSGV-LPLSKTNIKNLAVIGPNANATTVMISNYA 435
GP DVCT+ +Q LALEAA+QGIVLL+NSG LPLS + LA+IGPN+NAT MI NYA
Sbjct: 22 GPRDVCTQPHQELALEAARQGIVLLKNSGPSLPLSTRRHRTLAIIGPNSNATVTMIGNYA 201
Query: 436 GIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSI 495
GI RYTSP +G+ KY ++ + GC NV D F +A AA ADA VLV+GLDQSI
Sbjct: 202 GIGYRYTSP*EGIGKYAKTI-HELGCANVAGTDDKQFGSALNAARQADATVLVMGLDQSI 378
Query: 496 EAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKS 543
EAE +DR L LPG Q+ LV VA A+KG ILV+M+ GP+DI+F K+
Sbjct: 379 EAETVDRAGLLLPGRQQDLVSKVAAASKGPTILVLMSGGPVDITFAKN 522
>TC19191 similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragment),
partial (31%)
Length = 770
Score = 176 bits (445), Expect = 2e-44
Identities = 90/208 (43%), Positives = 127/208 (60%)
Frame = +2
Query: 557 GQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNG 616
G+AGG AIA VIFG +NP GR P TWYPQSY D+VPMT+MNMR + + +PGRTYRFY G
Sbjct: 2 GEAGGAAIADVIFGFHNPSGRLPMTWYPQSYVDKVPMTNMNMRPDPTTGYPGRTYRFYKG 181
Query: 617 NSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSDGQAIDISTISCLD 676
+++ FG GLSYST + AP + S+ +++ SL +++D++ C +
Sbjct: 182 ETVFSFGDGLSYSTIEHKLVKAPQLV------SIPLAEDHVCHSLK-CKSLDVAGEHCEN 340
Query: 677 LTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLE 736
L F + + +KN G + H V +F PP V AP K L+GFE+ H+ V +
Sbjct: 341 LEFEIHLSIKNKGRMSSDHTVFLFSSPPA---VHNAPHKHLVGFEKVHLEGNTEALVKFK 511
Query: 737 IDICQLLSNVDSDGKRKLIIGKHTLLVG 764
+D+C+ LS VD G RK+ +G+H L VG
Sbjct: 512 VDVCKDLSVVDELGNRKVALGQHLLHVG 595
>CN825311
Length = 522
Score = 174 bits (441), Expect = 5e-44
Identities = 81/155 (52%), Positives = 107/155 (68%)
Frame = +2
Query: 188 GLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKS 247
G D + ++ D L VS+CCKH+TAYD++ W R++F+A V++QDLEDTYQPPF+
Sbjct: 59 GSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNFNAVVSQQDLEDTYQPPFRG 238
Query: 248 CVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYT 307
CV +G S +MCSYN VNG+P CA DLL GV R WG GYI SDCD+V +E Y
Sbjct: 239 CVQQGKASCLMCSYNEVNGVPACASEDLL-GVARNNWGFKGYITSDCDAVATVFEYQVYV 415
Query: 308 ATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKK 342
+ EDAVA LKAG ++NCG Y+ ++TA+AV+ K
Sbjct: 416 KSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQGK 520
>BP033690
Length = 546
Score = 158 bits (400), Expect = 3e-39
Identities = 86/157 (54%), Positives = 111/157 (69%), Gaps = 6/157 (3%)
Frame = +2
Query: 4 NLLF---LSLFLSLLVLLPIL--TSSQKHACNKA-SPKTSNFPFCDTTLSYEARAKDLVS 57
N LF L+LF + L ++ ++ AC+ A +P S + FC+ +L AR DLV
Sbjct: 74 NALFALPLTLFTTXLFSCGLVWGQTTPNFACDVAKNPALSGYGFCNRSLPVNARVADLVG 253
Query: 58 RLTLQEKAQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVIL 117
RLTLQEK LV+ ++ ++RLG+P YEWWSEALHGVSN+GPGTRF+ VP A SFP IL
Sbjct: 254 RLTLQEKIGNLVSGAAIVSRLGIPKYEWWSEALHGVSNIGPGTRFSNVVPRAASFPMPIL 433
Query: 118 SAASFNATLWYNMGQVVSTEARAMYNVDLAGLTFWSP 154
+AASFN +L+ +G+VVSTEARAMYNV LAGLT+ SP
Sbjct: 434 TAASFNTSLFQAIGKVVSTEARAMYNVGLAGLTYGSP 544
>TC18444 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial (12%)
Length = 387
Score = 132 bits (333), Expect = 2e-31
Identities = 59/86 (68%), Positives = 70/86 (80%)
Frame = +2
Query: 547 IGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNF 606
IGGILW GYPG+ GG A+AQVIFG++NPGGR P TWYP+ Y +VPMTDM MRA+ + +
Sbjct: 5 IGGILWAGYPGELGGIALAQVIFGDHNPGGRLPITWYPKDYI-KVPMTDMRMRADPATGY 181
Query: 607 PGRTYRFYNGNSLYEFGHGLSYSTFS 632
PGRTYRFY G LYEFG+GLSYS +S
Sbjct: 182 PGRTYRFYKGPKLYEFGYGLSYSKYS 259
>AV775420
Length = 457
Score = 131 bits (329), Expect = 5e-31
Identities = 71/147 (48%), Positives = 102/147 (69%), Gaps = 3/147 (2%)
Frame = +1
Query: 314 VALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFFD-NPKSLP 372
VA L+AG+++ CGDYL K+ +AV KKV +S +D+AL + + +RLG FD NP L
Sbjct: 19 VADVLRAGMDVECGDYLTKHGKSAV*QKKVPISQIDRALHNLFSIRIRLGLFDGNPSKLT 198
Query: 373 FANLGPSDVCTKENQLLALEAAKQGIVLLENSG-VLPLSKTNIKNLAVIGPNANATTV-M 430
+ ++GP+ VC+K+N LALEAA+ GIVLL+ + +LPL KTN ++AVIGPNANA+++ +
Sbjct: 199 YGSIGPNQVCSKQNLKLALEAARNGIVLLKKTASILPLPKTN-PSMAVIGPNANASSLAV 375
Query: 431 ISNYAGIPCRYTSPLQGLQKYISSVTY 457
+ NY G PC+ + LQG Q Y Y
Sbjct: 376 LGNYYGRPCKLVTLLQGFQHYAKDTIY 456
>AV415013
Length = 154
Score = 108 bits (271), Expect = 3e-24
Identities = 51/51 (100%), Positives = 51/51 (100%)
Frame = +2
Query: 530 IMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPF 580
IMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPF
Sbjct: 2 IMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPF 154
>TC13306 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (12%)
Length = 435
Score = 108 bits (270), Expect = 3e-24
Identities = 51/95 (53%), Positives = 63/95 (65%)
Frame = +2
Query: 28 ACNKASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEKAQQLVNPSSGIARLGVPSYEWWS 87
AC+ T F FC T + R +DL+ RLTL EK + +VN + + RLG+ YEWWS
Sbjct: 149 ACDPQKGLTRGFKFCRTNVPIHVRVQDLIGRLTLAEKIRLVVNNAIEVPRLGIQGYEWWS 328
Query: 88 EALHGVSNLGPGTRFNRTVPGATSFPAVILSAASF 122
EALHGVSN+GPGT+F P ATSFP VI +AASF
Sbjct: 329 EALHGVSNVGPGTKFGGAFPAATSFPQVITTAASF 433
>AV427695
Length = 202
Score = 77.0 bits (188), Expect = 1e-14
Identities = 41/67 (61%), Positives = 48/67 (71%)
Frame = +2
Query: 472 FAAATKAAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIM 531
F A AA ADA VLV+GLDQSIEAE DRV L LPG Q+ LV VA A++G ILV+M
Sbjct: 2 FGPALDAARHADATVLVMGLDQSIEAETRDRVGLLLPGHQQDLVSKVAAASRGPTILVLM 181
Query: 532 AAGPIDI 538
+ GP+DI
Sbjct: 182 SGGPLDI 202
>TC19733 weakly similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial
(15%)
Length = 598
Score = 72.0 bits (175), Expect = 3e-13
Identities = 43/133 (32%), Positives = 69/133 (51%)
Frame = +2
Query: 654 HNNIFESLSDGQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAP 713
H + + +AI ++ C L+ L + VKN G +G+H +LVF PP A
Sbjct: 29 HRHANSTNISNKAIRVTHARCGKLSIALHVNVKNVGSRDGTHTLLVFSAPPDGG-AHWAV 205
Query: 714 IKQLIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRH 773
KQL+ FE+ HV + V + I +C+LLS VD G R++ +G+H+L +G V+H
Sbjct: 206 QKQLVAFEKVHVPAKSQQRVQINIHVCKLLSVVDRSGIRRIPMGEHSLHIG-----DVKH 370
Query: 774 QIDISLSGNGMEK 786
+ + G+ K
Sbjct: 371 SVSLQADAIGIIK 409
>TC15933 weakly similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragment),
partial (14%)
Length = 453
Score = 58.2 bits (139), Expect = 5e-09
Identities = 32/81 (39%), Positives = 45/81 (55%)
Frame = +1
Query: 685 VKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLS 744
VKN G SH VL+F+ PP V AP K L+G E+ H+ V +D+C+ LS
Sbjct: 13 VKNMGKMTSSHTVLLFFTPPA---VHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLS 183
Query: 745 NVDSDGKRKLIIGKHTLLVGS 765
VD R++ +G+H L VG+
Sbjct: 184 VVDELAHRRVPLGQHLLHVGN 246
>AV780323
Length = 544
Score = 44.3 bits (103), Expect = 8e-05
Identities = 22/72 (30%), Positives = 37/72 (50%)
Frame = -3
Query: 715 KQLIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQ 774
KQL+ FE+ H+ V + I +C+LLS VD G R++ +G+H +G + H
Sbjct: 539 KQLVAFEKVHIPAKAQRRVRVNIHVCKLLSVVDRSGIRRIPMGEHRFQIG-----DIEHS 375
Query: 775 IDISLSGNGMEK 786
+ + G+ K
Sbjct: 374 LSLQAKTLGVIK 339
>TC15428
Length = 629
Score = 38.1 bits (87), Expect = 0.005
Identities = 18/48 (37%), Positives = 26/48 (53%)
Frame = +1
Query: 673 SCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGF 720
+C ++ + +GVKN G G H VL+F P G P+K L+GF
Sbjct: 130 TCQSMSVSVTLGVKNDGSMAGKHPVLLFMRPGKQR--NGNPLKPLVGF 267
>TC12358 weakly similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (6%)
Length = 395
Score = 33.9 bits (76), Expect = 0.10
Identities = 16/28 (57%), Positives = 18/28 (64%)
Frame = +1
Query: 37 SNFPFCDTTLSYEARAKDLVSRLTLQEK 64
+ F FCD +L R DLV RLTLQEK
Sbjct: 301 AGFGFCDKSLPVADRVADLVKRLTLQEK 384
>BP075556
Length = 469
Score = 32.3 bits (72), Expect = 0.30
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 1/80 (1%)
Frame = -3
Query: 686 KNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGF-ERAHVMVGMTEFVTLEIDICQLLS 744
+N G + H + +F PP V P L G E + T ++D+C+ LS
Sbjct: 443 QNQGRMSSDHTLFLFSSPPP---VPTPPSSTLTGI*ESSLGRKYRTPLAKFKVDVCKDLS 273
Query: 745 NVDSDGKRKLIIGKHTLLVG 764
VD G ++ +G+H G
Sbjct: 272 VVDELGHSQVALGQHLASCG 213
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,923,139
Number of Sequences: 28460
Number of extensions: 169922
Number of successful extensions: 926
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 905
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 907
length of query: 794
length of database: 4,897,600
effective HSP length: 98
effective length of query: 696
effective length of database: 2,108,520
effective search space: 1467529920
effective search space used: 1467529920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0218.19


