

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.18
(330 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
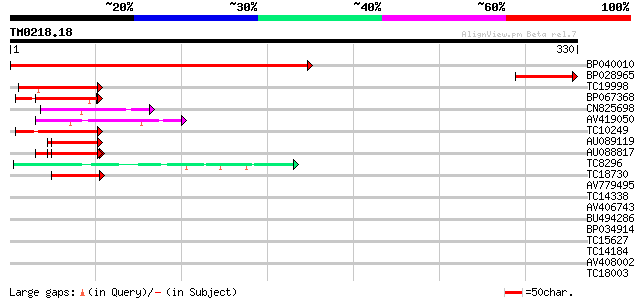
Score E
Sequences producing significant alignments: (bits) Value
BP040010 366 e-102
BP028965 78 2e-15
TC19998 similar to UP|RED_HUMAN (Q13123) Red protein (RER protei... 50 4e-07
BP067368 41 2e-04
CN825698 41 2e-04
AV419050 41 2e-04
TC10249 similar to UP|Q84LL9 (Q84LL9) Salt tolerance protein 3, ... 41 2e-04
AU089119 41 3e-04
AU088817 41 3e-04
TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1... 40 7e-04
TC18730 similar to UP|Q963J2 (Q963J2) Voltage-dependent calcium ... 40 7e-04
AV779495 39 0.001
TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%) 38 0.002
AV406743 38 0.003
BU494286 37 0.005
BP034914 36 0.010
TC15627 35 0.013
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 35 0.013
AV408002 34 0.030
TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 34 0.030
>BP040010
Length = 567
Score = 366 bits (939), Expect = e-102
Identities = 176/176 (100%), Positives = 176/176 (100%)
Frame = +2
Query: 1 MSHRDSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGG 60
MSHRDSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGG
Sbjct: 38 MSHRDSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGG 217
Query: 61 GGAGGDQNPTDRDQRNLPPPPHSKQESVGAGDKKSNDHQEPPKHSAQPPRSRSYYQHAER 120
GGAGGDQNPTDRDQRNLPPPPHSKQESVGAGDKKSNDHQEPPKHSAQPPRSRSYYQHAER
Sbjct: 218 GGAGGDQNPTDRDQRNLPPPPHSKQESVGAGDKKSNDHQEPPKHSAQPPRSRSYYQHAER 397
Query: 121 GNTEQVGRSAGRREAGGKAFAQSKENSERVETSQSREQRDEKSQAKLDDNLHRRDG 176
GNTEQVGRSAGRREAGGKAFAQSKENSERVETSQSREQRDEKSQAKLDDNLHRRDG
Sbjct: 398 GNTEQVGRSAGRREAGGKAFAQSKENSERVETSQSREQRDEKSQAKLDDNLHRRDG 565
Score = 31.2 bits (69), Expect = 0.25
Identities = 25/78 (32%), Positives = 34/78 (43%), Gaps = 10/78 (12%)
Frame = +2
Query: 218 SHTDHPPERNERKEERSGNPRHLERHEKQIAGDRAPNRSEGR-RDGFSSRARY------- 269
SH D + K +R +P+ R KQ DR +R R R+ S R R
Sbjct: 41 SHRDSDSKLRHSKFDREPSPKRYRRDGKQ---DRERDRDRDRERERGSERGRDRDRVTTN 211
Query: 270 --GGSGGNNNDRGRDKFN 285
GG+GG+ N RD+ N
Sbjct: 212 GGGGAGGDQNPTDRDQRN 265
>BP028965
Length = 448
Score = 77.8 bits (190), Expect = 2e-15
Identities = 36/36 (100%), Positives = 36/36 (100%)
Frame = -3
Query: 295 TRTEKWKHDLFQEADNDPVPKNEDDQIAKLEALLAS 330
TRTEKWKHDLFQEADNDPVPKNEDDQIAKLEALLAS
Sbjct: 446 TRTEKWKHDLFQEADNDPVPKNEDDQIAKLEALLAS 339
>TC19998 similar to UP|RED_HUMAN (Q13123) Red protein (RER protein) (IK
factor) (Cytokine IK), partial (6%)
Length = 492
Score = 50.4 bits (119), Expect = 4e-07
Identities = 25/52 (48%), Positives = 33/52 (63%), Gaps = 3/52 (5%)
Frame = +1
Query: 6 SDSKLRHSKF---DREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDR 54
+D + RH + PS RRD +DR+RDRDRDRER+R +R RDR+R
Sbjct: 256 ADQRRRHQQILSDSNPPSADPNRRDRDRDRDRDRDRDRERDRERDRDRDRER 411
Score = 34.3 bits (77), Expect = 0.030
Identities = 23/53 (43%), Positives = 33/53 (61%), Gaps = 3/53 (5%)
Frame = +1
Query: 6 SDSKLRHSKFDREPSPKRYR-RDGKQDRERDRD-RDRERERGSE-RGRDRDRV 55
+D R DR+ R R RD ++DR+RDR+ R+RERER E + R+R R+
Sbjct: 310 ADPNRRDRDRDRDRDRDRDRERDRERDRDRDRERRNRERERDEENKARERARL 468
>BP067368
Length = 322
Score = 41.2 bits (95), Expect = 2e-04
Identities = 20/46 (43%), Positives = 30/46 (64%), Gaps = 7/46 (15%)
Frame = -2
Query: 16 DREPSPKRYRRDGKQDRERDRDRDRERERG-------SERGRDRDR 54
DR+ R+ RD + R+RDRDR+RER+R ++RGR+R+R
Sbjct: 144 DRDHREDRHHRDRDRTRDRDRDRERERDRDVTEIEDVTDRGRERER 7
Score = 40.8 bits (94), Expect = 3e-04
Identities = 26/52 (50%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Frame = -2
Query: 4 RDSDSKLRHSKFDREPSPKRYR-RDGKQDRE-RDRDRDRERERGSERGRDRD 53
+D D + S RE S R R RD ++DR RDRDR R+R+R ER RDRD
Sbjct: 201 KDKDGERERS---RERSSDRARDRDHREDRHHRDRDRTRDRDRDRERERDRD 55
Score = 35.8 bits (81), Expect = 0.010
Identities = 20/52 (38%), Positives = 29/52 (55%)
Frame = -2
Query: 3 HRDSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDR 54
H+ + S+ S+ R + RD ++ RER +D+D ERER ER DR R
Sbjct: 291 HKSAPSR---SEEPRVREDRNVDRDREKSRERVKDKDGERERSRERSSDRAR 145
Score = 32.3 bits (72), Expect = 0.11
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 2/68 (2%)
Frame = -2
Query: 11 RHSKFDREPS-PKRYRRDG-KQDRERDRDRDRERERGSERGRDRDRVTTNGGGGAGGDQN 68
RHS ++ S P R ++DR DRDR++ RER ++ +R+R A +
Sbjct: 312 RHSGREQHKSAPSRSEEPRVREDRNVDRDREKSRERVKDKDGERERSRERSSDRARDRDH 133
Query: 69 PTDRDQRN 76
DR R+
Sbjct: 132 REDRHHRD 109
Score = 27.7 bits (60), Expect = 2.8
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 14/55 (25%)
Frame = -2
Query: 4 RDSD-SKLRHSKFDREPSPKRYRRDGKQDRERDRDRD-------------RERER 44
R SD ++ R + DR + RD +DRER+RDRD RER+R
Sbjct: 165 RSSDRARDRDHREDRHHRDRDRTRDRDRDRERERDRDVTEIEDVTDRGRERERDR 1
>CN825698
Length = 704
Score = 41.2 bits (95), Expect = 2e-04
Identities = 25/70 (35%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Frame = +1
Query: 19 PSPKRYRRDGKQDRERDRDRDR----ERERGSERGRDRDRVTTNGGGGAGGDQNPTDRDQ 74
P P RRD + DR+ DR +R +R S +DRDR + Q DRD+
Sbjct: 196 PPPPARRRDRRDDRDFDRPPNRRDYYDRSNNSPPPKDRDRDFKRRRSPSPPSQR--DRDR 369
Query: 75 RNLPPPPHSK 84
R+ PPPP+ +
Sbjct: 370 RHSPPPPYKR 399
Score = 29.6 bits (65), Expect = 0.73
Identities = 29/137 (21%), Positives = 47/137 (34%), Gaps = 4/137 (2%)
Frame = +1
Query: 158 QRDEKSQAKLDDNLHRRDGSPERKDDPPSNTRKRPAFREKKIQGDSGDANPAVSTERVK- 216
+RD + D +RRD + PP R R R + +P+ ++R +
Sbjct: 214 RRDRRDDRDFDRPPNRRDYYDRSNNSPPPKDRDRDFKRRR---------SPSPPSQRDRD 366
Query: 217 ASHTDHPPERNERKEERSGNPRHLERHEKQIAGDRAPNRSEGRRDGF---SSRARYGGSG 273
H+ PP + R+ G R+ R G R G+ S R+G
Sbjct: 367 RRHSPPPPYKRSRRGSPRGGYRYGPDDRFGYDNSGGYERGMGGRVGYVDDKSYGRFGHRS 546
Query: 274 GNNNDRGRDKFNARQGY 290
G + +GY
Sbjct: 547 AGEYQNGISDVESNRGY 597
>AV419050
Length = 409
Score = 41.2 bits (95), Expect = 2e-04
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 7/95 (7%)
Frame = +1
Query: 16 DREPSPKRYRRDGKQDRER---DRDRDRERERGSERGRDRDRVTTNGGGGAGGDQNPTDR 72
DR+ R RR K++R+R DRDRDR R S+R R R R +G ++P++R
Sbjct: 10 DRDRDRDRERRREKEERDRGDRDRDRDRTR---SKRSRSRSRSIDHGRSRHARSRSPSER 180
Query: 73 DQR----NLPPPPHSKQESVGAGDKKSNDHQEPPK 103
R P P ++ D +DH++ K
Sbjct: 181 SHRRRHHRTPSPDQPRKRH--RRDSVEDDHKDTKK 279
Score = 26.2 bits (56), Expect = 8.1
Identities = 26/126 (20%), Positives = 48/126 (37%)
Frame = +1
Query: 110 RSRSYYQHAERGNTEQVGRSAGRREAGGKAFAQSKENSERVETSQSREQRDEKSQAKLDD 169
R R + E+ ++ R R K +S+ S ++ +SR R +
Sbjct: 19 RDRDRERRREKEERDRGDRDRDRDRTRSK---RSRSRSRSIDHGRSRHARSRSPSERSHR 189
Query: 170 NLHRRDGSPERKDDPPSNTRKRPAFREKKIQGDSGDANPAVSTERVKASHTDHPPERNER 229
H R SP++ RKR R ++ D D T++V + D + ++
Sbjct: 190 RRHHRTPSPDQ-------PRKR--HRRDSVEDDHKD------TKKVVSDFVDGIVKEQQQ 324
Query: 230 KEERSG 235
K + +G
Sbjct: 325 KHKENG 342
>TC10249 similar to UP|Q84LL9 (Q84LL9) Salt tolerance protein 3, partial
(35%)
Length = 1096
Score = 41.2 bits (95), Expect = 2e-04
Identities = 21/51 (41%), Positives = 31/51 (60%)
Frame = +1
Query: 4 RDSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDR 54
R+ +++ R + +E +R RR DRE+ RD+DR+RER R RD DR
Sbjct: 172 REREAEERRKR--KEQEYERKRRGDSSDREKHRDKDRDRERDRYRDRDSDR 318
Score = 35.8 bits (81), Expect = 0.010
Identities = 24/74 (32%), Positives = 32/74 (42%)
Frame = +1
Query: 5 DSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGGGGAG 64
DS + +H DR+ RYR DR+ DR+R RER+ R R + G G
Sbjct: 238 DSSDREKHRDKDRDRERDRYR-----DRDSDRERSRERDVRGYRDGGRGVDSRLGNRRNG 402
Query: 65 GDQNPTDRDQRNLP 78
G DR + P
Sbjct: 403 GRDRYRDRSRSRSP 444
>AU089119
Length = 113
Score = 40.8 bits (94), Expect = 3e-04
Identities = 17/30 (56%), Positives = 26/30 (86%)
Frame = +2
Query: 25 RRDGKQDRERDRDRDRERERGSERGRDRDR 54
RR+ +++RER+R+R+RERER ER R+R+R
Sbjct: 2 RRERERERERERERERERERERERERERER 91
Score = 40.0 bits (92), Expect = 5e-04
Identities = 17/32 (53%), Positives = 26/32 (81%)
Frame = +2
Query: 23 RYRRDGKQDRERDRDRDRERERGSERGRDRDR 54
R R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 2 RRERERERERERERERERERERERERERERER 97
Score = 38.9 bits (89), Expect = 0.001
Identities = 16/29 (55%), Positives = 25/29 (86%)
Frame = +3
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDR 54
R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 21 RERERERERERERERERERERERERERER 107
Score = 38.9 bits (89), Expect = 0.001
Identities = 16/29 (55%), Positives = 25/29 (86%)
Frame = +2
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDR 54
R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 17 RERERERERERERERERERERERERERER 103
Score = 38.9 bits (89), Expect = 0.001
Identities = 16/29 (55%), Positives = 25/29 (86%)
Frame = +1
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDR 54
R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 25 RERERERERERERERERERERERERERER 111
Score = 38.9 bits (89), Expect = 0.001
Identities = 16/29 (55%), Positives = 25/29 (86%)
Frame = +2
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDR 54
R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 23 RERERERERERERERERERERERERERER 109
Score = 38.9 bits (89), Expect = 0.001
Identities = 16/29 (55%), Positives = 25/29 (86%)
Frame = +3
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDR 54
R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 15 RERERERERERERERERERERERERERER 101
Score = 37.7 bits (86), Expect = 0.003
Identities = 16/37 (43%), Positives = 26/37 (70%)
Frame = +2
Query: 17 REPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRD 53
R + R+ +++RER+R+R+RERER ER R+R+
Sbjct: 2 RRERERERERERERERERERERERERERERERERERE 112
>AU088817
Length = 191
Score = 40.8 bits (94), Expect = 3e-04
Identities = 17/30 (56%), Positives = 26/30 (86%)
Frame = +1
Query: 25 RRDGKQDRERDRDRDRERERGSERGRDRDR 54
RR+ +++RER+R+R+RERER ER R+R+R
Sbjct: 4 RRERERERERERERERERERERERERERER 93
Score = 40.4 bits (93), Expect = 4e-04
Identities = 20/41 (48%), Positives = 31/41 (74%), Gaps = 1/41 (2%)
Frame = +2
Query: 16 DREPSPKRYR-RDGKQDRERDRDRDRERERGSERGRDRDRV 55
+RE +R R R+ +++RER+R+R+RERER ER R+R+ V
Sbjct: 14 EREREREREREREREREREREREREREREREXEREREREXV 136
Score = 40.0 bits (92), Expect = 5e-04
Identities = 17/32 (53%), Positives = 26/32 (81%)
Frame = +1
Query: 23 RYRRDGKQDRERDRDRDRERERGSERGRDRDR 54
R R+ +++RER+R+R+RERER ER R+R+R
Sbjct: 4 RRERERERERERERERERERERERERERERER 99
Score = 39.3 bits (90), Expect = 0.001
Identities = 16/35 (45%), Positives = 26/35 (73%)
Frame = +2
Query: 19 PSPKRYRRDGKQDRERDRDRDRERERGSERGRDRD 53
P + R+ +++RER+R+R+RERER ER R+R+
Sbjct: 2 PGERERERERERERERERERERERERERERERERE 106
Score = 39.3 bits (90), Expect = 0.001
Identities = 21/53 (39%), Positives = 29/53 (54%)
Frame = +3
Query: 30 QDRERDRDRDRERERGSERGRDRDRVTTNGGGGAGGDQNPTDRDQRNLPPPPH 82
Q+RER+R+R+RERER ER R+R+R + +R PPPH
Sbjct: 3 QERERERERERERERERERERERERERERERERERXRERERERXFCLSIPPPH 161
Score = 38.5 bits (88), Expect = 0.002
Identities = 19/40 (47%), Positives = 30/40 (74%), Gaps = 1/40 (2%)
Frame = +3
Query: 16 DREPSPKRYR-RDGKQDRERDRDRDRERERGSERGRDRDR 54
+RE +R R R+ +++RER+R+R+RERER R R+R+R
Sbjct: 12 ERERERERERERERERERERERERERERERERXRERERER 131
Score = 38.5 bits (88), Expect = 0.002
Identities = 17/38 (44%), Positives = 26/38 (67%)
Frame = +1
Query: 17 REPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDR 54
R + R+ +++RER+R+R+RERER ER R R+R
Sbjct: 4 RRERERERERERERERERERERERERERERERERXRER 117
Score = 38.5 bits (88), Expect = 0.002
Identities = 19/40 (47%), Positives = 30/40 (74%), Gaps = 1/40 (2%)
Frame = +2
Query: 16 DREPSPKRYR-RDGKQDRERDRDRDRERERGSERGRDRDR 54
+RE +R R R+ +++RER+R+R+RERER E R+R+R
Sbjct: 8 EREREREREREREREREREREREREREREREREXERERER 127
Score = 32.3 bits (72), Expect = 0.11
Identities = 17/37 (45%), Positives = 26/37 (69%), Gaps = 1/37 (2%)
Frame = +1
Query: 11 RHSKFDREPSPKRYR-RDGKQDRERDRDRDRERERGS 46
R + +RE +R R R+ +++RER+R R+RERER S
Sbjct: 25 RERERERERERERERERERERERERERXRERERERXS 135
>TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;}, partial (55%)
Length = 1276
Score = 39.7 bits (91), Expect = 7e-04
Identities = 52/179 (29%), Positives = 66/179 (36%), Gaps = 13/179 (7%)
Frame = +2
Query: 3 HRDSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGGGG 62
HRD R + R + Y + + DR R RDRD R R R R G G
Sbjct: 461 HRDDHRSSRDRDYRRRSRSRSYDKYERGDRHRGRDRDYRR-----RSRSRSASLDYKGRG 625
Query: 63 AGGDQNPTDRDQRNLPPPPHSKQESVGAGDKKSNDHQEP-------PKHSAQPPRSRSYY 115
G R P S+ SV D +S + P PK S P RS S
Sbjct: 626 RG----------RYDDEPSRSRSRSV---DSRSPARRSPSPRRSPSPKRSVSPKRSTS-P 763
Query: 116 QHAERG----NTEQVGRSAGRREAG--GKAFAQSKENSERVETSQSREQRDEKSQAKLD 168
+ + RG N + GRS R G+ A S+ S R +EK Q +LD
Sbjct: 764 RKSPRGESPDNRSRDGRSPTPRSVSPRGRPVA-SRSPSPRKSNGDE*TVFNEKFQRRLD 937
>TC18730 similar to UP|Q963J2 (Q963J2) Voltage-dependent calcium channel
alpha13 subunit (Fragment), partial (5%)
Length = 515
Score = 39.7 bits (91), Expect = 7e-04
Identities = 17/31 (54%), Positives = 26/31 (83%)
Frame = +1
Query: 25 RRDGKQDRERDRDRDRERERGSERGRDRDRV 55
RR+ +++RER+R+R+RERER ER R+R+ V
Sbjct: 7 RRERERERERERERERERERERERERERELV 99
Score = 38.1 bits (87), Expect = 0.002
Identities = 16/35 (45%), Positives = 26/35 (73%)
Frame = +3
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDRVTTNGG 60
++ +++RER+R+R+RERER ER R+R+R G
Sbjct: 6 QERERERERERERERERERERERERERERARAEFG 110
Score = 35.4 bits (80), Expect = 0.013
Identities = 15/29 (51%), Positives = 23/29 (78%)
Frame = +1
Query: 23 RYRRDGKQDRERDRDRDRERERGSERGRD 51
R R+ +++RER+R+R+RERER ER R+
Sbjct: 7 RRERERERERERERERERERERERERERE 93
Score = 35.0 bits (79), Expect = 0.017
Identities = 16/33 (48%), Positives = 24/33 (72%)
Frame = +3
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDRVTTN 58
R+ +++RER+R+R+RERER ER R R T+
Sbjct: 18 RERERERERERERERERERERERERARAEFGTS 116
Score = 33.9 bits (76), Expect = 0.039
Identities = 14/29 (48%), Positives = 22/29 (75%)
Frame = +2
Query: 20 SPKRYRRDGKQDRERDRDRDRERERGSER 48
+P R+ +++RER+R+R+RERER ER
Sbjct: 2 APGERERERERERERERERERERERERER 88
Score = 32.7 bits (73), Expect = 0.086
Identities = 13/29 (44%), Positives = 21/29 (71%)
Frame = +2
Query: 19 PSPKRYRRDGKQDRERDRDRDRERERGSE 47
P + R+ +++RER+R+R+RERER E
Sbjct: 5 PGERERERERERERERERERERERERERE 91
Score = 30.4 bits (67), Expect = 0.43
Identities = 12/28 (42%), Positives = 20/28 (70%)
Frame = +1
Query: 17 REPSPKRYRRDGKQDRERDRDRDRERER 44
R + R+ +++RER+R+R+RERER
Sbjct: 7 RRERERERERERERERERERERERERER 90
>AV779495
Length = 480
Score = 38.9 bits (89), Expect = 0.001
Identities = 17/25 (68%), Positives = 22/25 (88%)
Frame = -3
Query: 30 QDRERDRDRDRERERGSERGRDRDR 54
+DRERDR+R+RERE +ER RDR+R
Sbjct: 436 RDRERDREREREREIHTERQRDRER 362
Score = 35.0 bits (79), Expect = 0.017
Identities = 15/29 (51%), Positives = 22/29 (75%)
Frame = -3
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDR 54
RD ++DRER+R+R+ ER +R RDR+R
Sbjct: 436 RDRERDREREREREIHTERQRDRERDRER 350
Score = 31.2 bits (69), Expect = 0.25
Identities = 20/39 (51%), Positives = 25/39 (63%), Gaps = 1/39 (2%)
Frame = -2
Query: 17 REPSPKRYR-RDGKQDRERDRDRDRERERGSERGRDRDR 54
RE RYR RDG + RER R+R+RER+ E R R+R
Sbjct: 476 RERDI*RYRDRDG*R*RER*RERERERDTHRETER*RER 360
Score = 30.4 bits (67), Expect = 0.43
Identities = 16/30 (53%), Positives = 21/30 (69%), Gaps = 1/30 (3%)
Frame = -3
Query: 16 DREPSPKRYR-RDGKQDRERDRDRDRERER 44
DRE +R R R+ +R+RDR+RDRER R
Sbjct: 433 DRERDREREREREIHTERQRDRERDRERAR 344
Score = 26.9 bits (58), Expect = 4.7
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = -3
Query: 32 RERDRDRDRERERGSERGRDRDR 54
RER+ RD E E G +R RDR+R
Sbjct: 478 REREIYRDIEIEMGRDRERDRER 410
>TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%)
Length = 1751
Score = 38.1 bits (87), Expect = 0.002
Identities = 26/87 (29%), Positives = 36/87 (40%)
Frame = +2
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDRVTTNGGGGAGGDQNPTDRDQRNLPPPPHSKQ 85
R+ + ER+R DR R R DR R +N +RD +N PPP
Sbjct: 644 RNNARANERNRRNDRPRNNDRSRNFDRRR------------ENVVNRDYQNRPPP----- 772
Query: 86 ESVGAGDKKSNDHQEPPKHSAQPPRSR 112
AG SN PP ++ PP ++
Sbjct: 773 ---NAGGPPSNQGGFPPNNAGGPPSNQ 844
>AV406743
Length = 365
Score = 37.7 bits (86), Expect = 0.003
Identities = 20/56 (35%), Positives = 33/56 (58%), Gaps = 4/56 (7%)
Frame = +3
Query: 31 DRERDRDRDRERERGSERGRDRDRVTTNGGGGAGGDQNPTDRDQRNLP----PPPH 82
+RER+R+R+RERER ER R+R+R G + + +R++ ++ P PH
Sbjct: 3 ERERERERERERERERERERERER------GDSSSSRGREERERMSIAFLSVPTPH 152
Score = 37.4 bits (85), Expect = 0.003
Identities = 17/34 (50%), Positives = 25/34 (73%)
Frame = +2
Query: 29 KQDRERDRDRDRERERGSERGRDRDRVTTNGGGG 62
+++RER+R+R+RERER ER R+R + G GG
Sbjct: 5 ERERERERERERERERERERERERG*LKLKG*GG 106
Score = 37.0 bits (84), Expect = 0.005
Identities = 16/34 (47%), Positives = 25/34 (73%)
Frame = +1
Query: 29 KQDRERDRDRDRERERGSERGRDRDRVTTNGGGG 62
+++RER+R+R+RERER ER R+R+ + G G
Sbjct: 1 EREREREREREREREREREREREREGIAQAQGVG 102
Score = 34.7 bits (78), Expect = 0.023
Identities = 14/24 (58%), Positives = 21/24 (87%)
Frame = +2
Query: 26 RDGKQDRERDRDRDRERERGSERG 49
R+ +++RER+R+R+RERER ERG
Sbjct: 8 RERERERERERERERERERERERG 79
Score = 34.7 bits (78), Expect = 0.023
Identities = 14/24 (58%), Positives = 21/24 (87%)
Frame = +3
Query: 26 RDGKQDRERDRDRDRERERGSERG 49
R+ +++RER+R+R+RERER ERG
Sbjct: 6 RERERERERERERERERERERERG 77
>BU494286
Length = 198
Score = 37.0 bits (84), Expect = 0.005
Identities = 17/37 (45%), Positives = 20/37 (53%)
Frame = +3
Query: 26 RDGKQDRERDRDRDRERERGSERGRDRDRVTTNGGGG 62
R+G++ RDRDRDR R R R R R R GG
Sbjct: 36 REGRERERRDRDRDRGRRRSRSRSRSRSRDRKEHDGG 146
Score = 32.7 bits (73), Expect = 0.086
Identities = 20/51 (39%), Positives = 28/51 (54%)
Frame = +3
Query: 10 LRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGG 60
+RH + D + R+ ++DR+RDR R R R R R R RDR +GG
Sbjct: 3 IRHEEKDYGRDREGRERE-RRDRDRDRGRRRSRSR--SRSRSRDRKEHDGG 146
>BP034914
Length = 544
Score = 35.8 bits (81), Expect = 0.010
Identities = 27/79 (34%), Positives = 36/79 (45%)
Frame = +3
Query: 5 DSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERGSERGRDRDRVTTNGGGGAG 64
DSDS S R +R R DG RER+ R RE+ + RDRDR T +
Sbjct: 105 DSDSDSDRSSRRRREKERRRRSDGGSKRERESSRRREK----RKKRDRDRKKTKRHRHS- 269
Query: 65 GDQNPTDRDQRNLPPPPHS 83
D++ + D+ L P S
Sbjct: 270 DDESHSSEDEDKLRVQPLS 326
Score = 32.0 bits (71), Expect = 0.15
Identities = 26/107 (24%), Positives = 44/107 (40%)
Frame = +3
Query: 65 GDQNPTDRDQRNLPPPPHSKQESVGAGDKKSNDHQEPPKHSAQPPRSRSYYQHAERGNTE 124
GD+ R + P P S S A D S+ + + + R RS G ++
Sbjct: 24 GDKRKKQRYVHDDTPSPSSSSSSSTASDSDSDSDRSSRRRREKERRRRS------DGGSK 185
Query: 125 QVGRSAGRREAGGKAFAQSKENSERVETSQSREQRDEKSQAKLDDNL 171
+ S+ RRE + K + +R +T + R DE ++ +D L
Sbjct: 186 RERESSRRRE------KRKKRDRDRKKTKRHRHSDDESHSSEDEDKL 308
>TC15627
Length = 725
Score = 35.4 bits (80), Expect = 0.013
Identities = 28/103 (27%), Positives = 40/103 (38%), Gaps = 12/103 (11%)
Frame = -3
Query: 151 ETSQSREQRDEKSQAKLDDNLHRRDGSPERKDDPPSNTRKRPAFREKKIQGDSGDANPAV 210
E + S L D R +P +P + RKRP KK++G D +
Sbjct: 507 EIKRKNNNTSISSTNSLADRTRRNKPNP----NPKTEKRKRP----KKLKGTQEDGEDLM 352
Query: 211 ST------------ERVKASHTDHPPERNERKEERSGNPRHLE 241
T + ++ T +R KEE +GNPRHLE
Sbjct: 351 LTKITTPITFFKMFQTIRGGSTMERNQRRA*KEESNGNPRHLE 223
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 35.4 bits (80), Expect = 0.013
Identities = 35/126 (27%), Positives = 53/126 (41%), Gaps = 3/126 (2%)
Frame = +3
Query: 57 TNGGGGAGGDQNPTDRDQRNLPPPP--HSKQESVG-AGDKKSNDHQEPPKHSAQPPRSRS 113
+ GGGG G D + RD+ + H K E +G G + + KH +S
Sbjct: 480 STGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKH-----KSEK 644
Query: 114 YYQHAERGNTEQVGRSAGRREAGGKAFAQSKENSERVETSQSREQRDEKSQAKLDDNLHR 173
++A R E+ +A AGG AF + E E ++ +E+S K +L
Sbjct: 645 DPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKE-------AKEEEEESHGKKHHHLFG 803
Query: 174 RDGSPE 179
RD S E
Sbjct: 804 RD*SIE 821
>AV408002
Length = 408
Score = 34.3 bits (77), Expect = 0.030
Identities = 13/24 (54%), Positives = 21/24 (87%)
Frame = -3
Query: 25 RRDGKQDRERDRDRDRERERGSER 48
R +G+++RER+R+R+RER+ G ER
Sbjct: 88 REEGERERERERERERERKVGGER 17
Score = 31.6 bits (70), Expect = 0.19
Identities = 12/25 (48%), Positives = 20/25 (80%)
Frame = -1
Query: 29 KQDRERDRDRDRERERGSERGRDRD 53
++ RER+R+R+RERER E+ +R+
Sbjct: 90 REKREREREREREREREREKWEERE 16
Score = 30.8 bits (68), Expect = 0.33
Identities = 13/26 (50%), Positives = 22/26 (84%), Gaps = 1/26 (3%)
Frame = -1
Query: 24 YRRDGKQ-DRERDRDRDRERERGSER 48
+RR+ ++ +RER+R+R+RERE+ ER
Sbjct: 96 FRREKREREREREREREREREKWEER 19
Score = 29.6 bits (65), Expect = 0.73
Identities = 12/26 (46%), Positives = 19/26 (72%)
Frame = -2
Query: 27 DGKQDRERDRDRDRERERGSERGRDR 52
D ++ RER+R+R+RERER R++
Sbjct: 92 DERRGREREREREREREREKSGRREK 15
Score = 26.9 bits (58), Expect = 4.7
Identities = 12/41 (29%), Positives = 24/41 (58%)
Frame = -1
Query: 5 DSDSKLRHSKFDREPSPKRYRRDGKQDRERDRDRDRERERG 45
D + R+S++ + R+ +++RER+R++ ERE G
Sbjct: 132 DGGEEGRYSEYLFRREKREREREREREREREREKWEERESG 10
>TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (42%)
Length = 587
Score = 34.3 bits (77), Expect = 0.030
Identities = 32/119 (26%), Positives = 52/119 (42%), Gaps = 18/119 (15%)
Frame = +1
Query: 29 KQDRERDRDRDRERERGSER---GRDRDRV-----TTNGGGGAGGDQNPTDRDQR--NLP 78
+++RER+R+R+RERER + G++ RV + GGGG GG + D +
Sbjct: 1 ERERERERERERERERDAIHALDGKNGWRVELSHNSKGGGGGRGGGRGRGGEDLKCYECG 180
Query: 79 PPPHSKQE--------SVGAGDKKSNDHQEPPKHSAQPPRSRSYYQHAERGNTEQVGRS 129
P H +E +G+G ++S P + P ++ RG RS
Sbjct: 181 EPGHFARECRSRGGSRGLGSGRRRS---PSPYRRRRSPSYGYGRRSYSPRGRRSPRPRS 348
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.307 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,396,135
Number of Sequences: 28460
Number of extensions: 85295
Number of successful extensions: 1664
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 1098
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1429
length of query: 330
length of database: 4,897,600
effective HSP length: 91
effective length of query: 239
effective length of database: 2,307,740
effective search space: 551549860
effective search space used: 551549860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0218.18


