

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
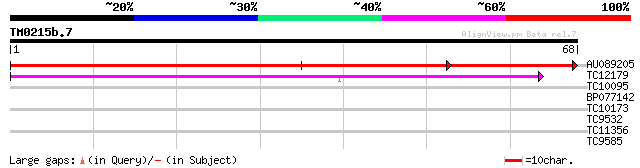
Query= TM0215b.7
(68 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU089205 102 1e-23
TC12179 weakly similar to UP|Q40709 (Q40709) WSI18 protein induc... 42 2e-05
TC10095 weakly similar to UP|Q94FU1 (Q94FU1) Auxin-regulated dua... 28 0.26
BP077142 25 2.2
TC10173 weakly similar to UP|Q9LJQ3 (Q9LJQ3) Lipid transfer prot... 25 2.8
TC9532 similar to UP|THIC_MYCLE (Q9ZBL0) Thiamine biosynthesis p... 25 2.8
TC11356 similar to GB|AAK44661.1|13879941|AE006947 thiamin biosy... 25 2.8
TC9585 similar to UP|O24078 (O24078) Protein phosphatase 2C, par... 24 6.3
>AU089205
Length = 396
Score = 102 bits (254), Expect = 1e-23
Identities = 52/53 (98%), Positives = 53/53 (99%)
Frame = +2
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGA 53
MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQG+
Sbjct: 38 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGS 196
Score = 37.0 bits (84), Expect = 7e-04
Identities = 18/33 (54%), Positives = 23/33 (69%)
Frame = +3
Query: 36 KETVQGAGQQIKASAQGAADAVKNATGMNNNSK 68
K+ +G + + + AADAVKNATGMNNNSK
Sbjct: 144 KKLCKGLVNRSRHQHRXAADAVKNATGMNNNSK 242
>TC12179 weakly similar to UP|Q40709 (Q40709) WSI18 protein induced by water
stress, partial (17%)
Length = 582
Score = 42.4 bits (98), Expect = 2e-05
Identities = 24/86 (27%), Positives = 40/86 (45%), Gaps = 22/86 (25%)
Frame = +2
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKET---------------------- 38
M S + ++NAGQ +GQ + K + M +A +A +
Sbjct: 65 MSSAQQNFNAGQNRGQTQAKAEDWMQSTKESASAAADRAHAAANTTGQTAEQNKEEAAGF 244
Query: 39 VQGAGQQIKASAQGAADAVKNATGMN 64
+Q G+Q+K AQGA ++VK+ GM+
Sbjct: 245 IQQTGEQVKNMAQGAVESVKHTLGMD 322
>TC10095 weakly similar to UP|Q94FU1 (Q94FU1) Auxin-regulated dual
specificity cytosolic kinase, partial (10%)
Length = 555
Score = 28.5 bits (62), Expect = 0.26
Identities = 15/46 (32%), Positives = 23/46 (49%)
Frame = +2
Query: 16 QAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+A E+ D A + V+G+G +AS +G ADA + AT
Sbjct: 155 KALEQLQEPKDTQKKGADPKGDRVRGSGLNHQASGKGGADATRKAT 292
>BP077142
Length = 385
Score = 25.4 bits (54), Expect = 2.2
Identities = 10/25 (40%), Positives = 16/25 (64%)
Frame = -3
Query: 44 QQIKASAQGAADAVKNATGMNNNSK 68
+++K AQGA +AVK+ GM +
Sbjct: 383 EKVKDMAQGATEAVKSTLGMGQKDE 309
>TC10173 weakly similar to UP|Q9LJQ3 (Q9LJQ3) Lipid transfer protein
(AT3g18280/MIE15_7), partial (77%)
Length = 671
Score = 25.0 bits (53), Expect = 2.8
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = +3
Query: 36 KETVQGAGQQIKASAQGAADAVKNAT 61
++ + G Q+ ASA GAA A+ + T
Sbjct: 501 RQGLNATGLQVTASAMGAASAISSTT 578
>TC9532 similar to UP|THIC_MYCLE (Q9ZBL0) Thiamine biosynthesis protein
thiC, partial (14%)
Length = 1259
Score = 25.0 bits (53), Expect = 2.8
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = +3
Query: 15 GQAEEKTSNLMDMASNAAQSAKETVQG 41
G AEE MD S QSAK+TV G
Sbjct: 216 GTAEEALLRGMDAMSAEFQSAKKTVSG 296
>TC11356 similar to GB|AAK44661.1|13879941|AE006947 thiamin biosynthesis
protein ThiC {Mycobacterium tuberculosis CDC1551;} ,
partial (15%)
Length = 565
Score = 25.0 bits (53), Expect = 2.8
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = +1
Query: 15 GQAEEKTSNLMDMASNAAQSAKETVQG 41
G AEE MD S QSAK+TV G
Sbjct: 253 GTAEEALLRGMDAMSAEFQSAKKTVSG 333
>TC9585 similar to UP|O24078 (O24078) Protein phosphatase 2C, partial (31%)
Length = 502
Score = 23.9 bits (50), Expect = 6.3
Identities = 12/46 (26%), Positives = 20/46 (43%)
Frame = -1
Query: 18 EEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATGM 63
EEK + E G++ +A A+ DAV++A G+
Sbjct: 328 EEKAVGGFGRRRRNGEEGDEEAAAEGEEAEAEAEAQDDAVEDALGL 191
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.295 0.108 0.262
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 370,183
Number of Sequences: 28460
Number of extensions: 1334
Number of successful extensions: 11
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10
length of query: 68
length of database: 4,897,600
effective HSP length: 44
effective length of query: 24
effective length of database: 3,645,360
effective search space: 87488640
effective search space used: 87488640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 48 (23.1 bits)
Lotus: description of TM0215b.7


