

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
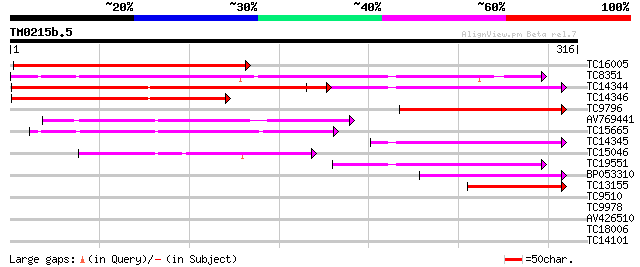
Query= TM0215b.5
(316 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16005 weakly similar to UP|Q96425 (Q96425) Polyketide reductas... 196 6e-51
TC8351 homologue to UP|Q41399 (Q41399) Chalcone reductase, complete 191 2e-49
TC14344 similar to UP|Q40310 (Q40310) Chalcone reductase, complete 171 1e-43
TC14346 similar to UP|Q40310 (Q40310) Chalcone reductase, partia... 134 2e-32
TC9796 similar to UP|Q8S4C1 (Q8S4C1) Chalcone reductase, partial... 106 6e-24
AV769441 101 2e-22
TC15665 similar to PIR|T02543|T02543 aldehyde dehydrogenase homo... 95 2e-20
TC14345 homologue to UP|Q39774 (Q39774) Polyketide reductase, pa... 88 2e-18
TC15046 similar to UP|Q9FVN7 (Q9FVN7) NADPH-dependent mannose 6-... 81 3e-16
TC19551 weakly similar to PIR|S57993|S57993 chalcone reductase h... 69 1e-12
BP053310 62 1e-10
TC13155 homologue to UP|Q9SXT0 (Q9SXT0) Chalcone reductase (Frag... 56 9e-09
TC9510 similar to UP|O22709 (O22709) F8A5.23 protein, partial (49%) 37 0.006
TC9978 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (36%) 36 0.010
AV426510 29 0.91
TC18006 weakly similar to UP|Q9M338 (Q9M338) Reductase-like prot... 28 1.6
TC14101 homologue to UP|Q39640 (Q39640) Glycolate oxidase , com... 26 7.7
>TC16005 weakly similar to UP|Q96425 (Q96425) Polyketide reductase (GGPKR1),
partial (40%)
Length = 474
Score = 196 bits (497), Expect = 6e-51
Identities = 100/132 (75%), Positives = 112/132 (84%)
Frame = +1
Query: 3 TTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQ 62
TTTVP+VVLQSSTG +R+ VMGL TAPEAA+ +ST D VLEA+KQG++HFDAAAAY VE+
Sbjct: 79 TTTVPQVVLQSSTGQRRMPVMGLGTAPEAASKVSTKDAVLEAIKQGYRHFDAAAAYGVEK 258
Query: 63 SVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHW 122
SVG AIAE Q LIAS+D LFVTSK+WV DNHP LIVPALQK LRTLQLEYLDLFLIHW
Sbjct: 259 SVGEAIAEALQLGLIASRDELFVTSKLWVTDNHPHLIVPALQKSLRTLQLEYLDLFLIHW 438
Query: 123 PFATKQGAVKYP 134
P T+ G VKYP
Sbjct: 439 PITTQPGGVKYP 474
>TC8351 homologue to UP|Q41399 (Q41399) Chalcone reductase, complete
Length = 1331
Score = 191 bits (484), Expect = 2e-49
Identities = 116/310 (37%), Positives = 183/310 (58%), Gaps = 11/310 (3%)
Frame = +1
Query: 1 MATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV 60
MA VP V+L S ++ MG S +N + + ++A++ G++HFD+A+ Y
Sbjct: 34 MAEKNVPNVLLNSGHK-MPVIGMGTSVDDRPSNDVLA-SIFVDAIQVGYRHFDSASVYGT 207
Query: 61 EQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLI 120
E+++G A+A+ LI S+D +F+TSK W D H +LIVPAL+ L+ L +EY+DL+LI
Sbjct: 208 EEAIGLALAKALDLGLIKSRDEVFITSKPWNTDAHQDLIVPALKTTLKKLGVEYVDLYLI 387
Query: 121 HWPFATK---QGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKL 177
HWP + + V + E +++ F ++G +MEE +LGL K+IG+ N KKL KL
Sbjct: 388 HWPVRLRHDLENPVVFSKE--DLLPFDIEGTWKAMEECYKLGLAKSIGICNFGTKKLTKL 561
Query: 178 FCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDN 236
ATI+P ++QVE++ WQQ KLR+FCK+K ++ + A+S L V+ G+ M+N
Sbjct: 562 LEIATITPAVNQVEMNPSWQQGKLREFCKQKGIH----VSAWSALGAYKVTWGSGAVMEN 729
Query: 237 DIQKELDT-HGKTIAQICL*WIYKH------VNFCDEEL*QGEDATKLAIFGCSLTENDF 289
I +++ T GKT+AQ+ L W+Y+ +F E + Q L IF L+E +
Sbjct: 730 HILQDIATAKGKTVAQVALRWVYQQGSSAMAKSFNKERMKQ-----NLEIFDFELSEEEL 894
Query: 290 EKISEIHQER 299
EKI ++ Q R
Sbjct: 895 EKIKQVPQRR 924
>TC14344 similar to UP|Q40310 (Q40310) Chalcone reductase, complete
Length = 1180
Score = 171 bits (434), Expect = 1e-43
Identities = 86/178 (48%), Positives = 123/178 (68%)
Frame = +2
Query: 2 ATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVE 61
A +P VL +++ +I V+G+ +AP+ T D ++EA+KQG++HFD AAAY E
Sbjct: 83 AAIEIPTRVLTNTSDQLKIPVIGMGSAPDFTCKKDTRDAIIEAIKQGYRHFDTAAAYGSE 262
Query: 62 QSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIH 121
Q++G A+ E L+ S++ LFVTSK+WV +NHP L+VPALQK L+TLQLEYLDL+LIH
Sbjct: 263 QALGEALKEALDLGLV-SREELFVTSKLWVTENHPHLVVPALQKSLKTLQLEYLDLYLIH 439
Query: 122 WPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFC 179
WP ++ G +PI + +++ F ++GV SMEE +LGLTKAIGV+N S+K C
Sbjct: 440 WPLSSTPGKFSFPIAMEDLLPFDVKGVWESMEECLKLGLTKAIGVSNFSVKTSKSALC 613
Score = 117 bits (294), Expect = 2e-27
Identities = 67/147 (45%), Positives = 89/147 (59%), Gaps = 2/147 (1%)
Frame = +3
Query: 166 VNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPLRMG 225
V+ S+ KL L A I P ++QVE++L WQQ +LR+FC + + AFSPLR G
Sbjct: 570 VSATSLLKLQNLLSVANILPAVNQVEMNLAWQQKELREFCNANGIV----LTAFSPLRKG 737
Query: 226 VSRGANLAMDNDIQKEL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCS 283
SRG N M+ND+ KE+ + HGKTIAQ+ L W+Y+ V F + + L IF +
Sbjct: 738 GSRGPNEVMENDMLKEIAEAHGKTIAQVSLRWLYEQGVTFAAKSYDKDRMNQNLQIFDWA 917
Query: 284 LTENDFEKISEIHQERLIKEPTKPLLN 310
LT+ D EKI +I Q RLI PTKP LN
Sbjct: 918 LTKEDLEKIDQIKQNRLIPGPTKPQLN 998
>TC14346 similar to UP|Q40310 (Q40310) Chalcone reductase, partial (38%)
Length = 435
Score = 134 bits (338), Expect = 2e-32
Identities = 67/122 (54%), Positives = 90/122 (72%)
Frame = +2
Query: 2 ATTTVPKVVLQSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVE 61
A +P VL +S+G QRI V+G+ +AP+ T D ++EA+KQG++HFD AAAY E
Sbjct: 68 AIIEIPTKVLTNSSGQQRIPVIGMGSAPDFTCKKDTRDAIIEAIKQGYRHFDTAAAYGSE 247
Query: 62 QSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIH 121
Q++G A+ E L+ S++ LFVTSK+WV +NHP L+VPALQK L+TLQLEYLDL+LIH
Sbjct: 248 QALGEALKEALDLGLV-SREELFVTSKLWVTENHPHLVVPALQKSLKTLQLEYLDLYLIH 424
Query: 122 WP 123
WP
Sbjct: 425 WP 430
>TC9796 similar to UP|Q8S4C1 (Q8S4C1) Chalcone reductase, partial (32%)
Length = 497
Score = 106 bits (264), Expect = 6e-24
Identities = 58/95 (61%), Positives = 68/95 (71%), Gaps = 2/95 (2%)
Frame = +2
Query: 218 AFSPLRMGVSRGANLAMDNDIQKEL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDAT 275
AFSPLR G S G NL MDNDI KEL D HG+TIAQICL W+Y+ + F + +
Sbjct: 2 AFSPLRKGASTGENLVMDNDILKELADAHGQTIAQICLRWLYEQGLTFVAKSYDKERMNQ 181
Query: 276 KLAIFGCSLTENDFEKISEIHQERLIKEPTKPLLN 310
L IF SLTE+D++KISEIHQ+RLIK PTKPLLN
Sbjct: 182 NLQIFDWSLTEDDYKKISEIHQDRLIKGPTKPLLN 286
>AV769441
Length = 594
Score = 101 bits (251), Expect = 2e-22
Identities = 59/174 (33%), Positives = 94/174 (53%)
Frame = +3
Query: 19 RILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIA 78
+I +GL T A +++ + + AV G++H D A Y E+ +G + + ++
Sbjct: 96 KIPSVGLGTWLAAPGVVA--NAIATAVDVGYRHLDCAQIYRNEKEIGDGLKKLFADGVVK 269
Query: 79 SKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVL 138
+D +F+TSK+W D+ PE + A + LR LQL+YLDL+LIHWP + K G + P
Sbjct: 270 RED-MFITSKLWCTDHLPEDVPKAFDRTLRDLQLDYLDLYLIHWPVSMKNGQLTKP---- 434
Query: 139 EIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEV 192
+ +ME G +AIGV+N S+KKL L A + P ++QVE+
Sbjct: 435 -----DIPSTWRAMEALYNSGKARAIGVSNFSVKKLQDLLEVANVPPAVNQVEL 581
>TC15665 similar to PIR|T02543|T02543 aldehyde dehydrogenase homolog
At2g37770 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (59%)
Length = 582
Score = 94.7 bits (234), Expect = 2e-20
Identities = 61/172 (35%), Positives = 97/172 (55%)
Frame = +1
Query: 12 QSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEE 71
Q +TG +I +GL T +++ V A++ G++H D A AY + +G+A+ +
Sbjct: 82 QLNTG-AKIPSVGLGTWQADPGVVAK--AVTTAIQVGYRHIDCAQAYNNQAEIGSALKKL 252
Query: 72 HQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAV 131
++ +D L++TSK+W D+ PE + A + L+ LQL+Y+DL+LIHWP TK+GAV
Sbjct: 253 FDDGVVKRED-LWITSKLWCSDHAPEDVPIAFDRTLQDLQLDYIDLYLIHWPIRTKKGAV 429
Query: 132 KYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATI 183
+ E L+ + + +ME G KAIGV+N S KKL L A +
Sbjct: 430 GFKPENLD--QPDIPSTWRAMEALFDSGKAKAIGVSNFSSKKLQDLLEIARV 579
>TC14345 homologue to UP|Q39774 (Q39774) Polyketide reductase, partial (35%)
Length = 516
Score = 87.8 bits (216), Expect = 2e-18
Identities = 51/111 (45%), Positives = 67/111 (59%), Gaps = 2/111 (1%)
Frame = +3
Query: 202 RDFCKEKALYWQFGIIAFSPLRMGVSRGANLAMDNDIQKEL-DTHGKTIAQICL*WIYKH 260
R+FC + + AFSPLR G SRGAN M+ND+ KE+ + HGK+IAQ+ L W+Y+
Sbjct: 3 REFCSANGIV----LTAFSPLRKGGSRGANEVMENDMLKEIAEAHGKSIAQVSLRWLYEQ 170
Query: 261 -VNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLIKEPTKPLLN 310
V F + + L IF +LT+ D EKI +I Q RLI PTKP LN
Sbjct: 171 GVTFAAKSYDKERMNQNLQIFDWALTKEDLEKIDQIKQNRLIPGPTKPQLN 323
>TC15046 similar to UP|Q9FVN7 (Q9FVN7) NADPH-dependent mannose 6-phosphate
reductase, partial (54%)
Length = 547
Score = 80.9 bits (198), Expect = 3e-16
Identities = 51/140 (36%), Positives = 81/140 (57%), Gaps = 7/140 (5%)
Frame = +1
Query: 39 DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPEL 98
D+VL ++K G++HFD AA Y E VG A+ E L+ +D LF+T+K+W D+
Sbjct: 130 DLVLNSIKLGYRHFDCAADYKNEPEVGEALKEAFDSGLVKRED-LFITTKLWNSDHGH-- 300
Query: 99 IVPALQKYLRTLQLEYLDLFLIHWPFATKQ------GAVKYPIEVLEI-MEFGMQGV*SS 151
++ A + L+ LQL+YLDL+L+H+P AT+ +VK VL+I ++ +
Sbjct: 301 VLEACKDSLKKLQLDYLDLYLVHFPVATRHTGVGTTDSVKGEDGVLDIDTTISLETTWHA 480
Query: 152 MEESQRLGLTKAIGVNNLSI 171
ME GL ++IG++N I
Sbjct: 481 MEGLVSSGLVRSIGISNYDI 540
>TC19551 weakly similar to PIR|S57993|S57993 chalcone reductase homolog -
Sesbania rostrata (fragment) {Sesbania rostrata;},
partial (81%)
Length = 624
Score = 68.9 bits (167), Expect = 1e-12
Identities = 47/122 (38%), Positives = 66/122 (53%), Gaps = 3/122 (2%)
Frame = +2
Query: 181 ATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQ 239
ATI P + QVE++ WQQ LR FCKEK ++ + A+SPL G G+ MD+ I
Sbjct: 8 ATIPPALVQVELNAAWQQENLRRFCKEKGIH----VSAWSPLGANGAMWGSLAVMDSPIL 175
Query: 240 KELD-THGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQ 297
K++ T KT+AQ+ L WI + + + L IF L+END EKI ++ Q
Sbjct: 176 KDIAITTEKTVAQVALRWIIEQGITPIVRSFNKERMKHNLEIFDWELSENDLEKIKQMPQ 355
Query: 298 ER 299
R
Sbjct: 356 HR 361
>BP053310
Length = 473
Score = 62.4 bits (150), Expect = 1e-10
Identities = 37/84 (44%), Positives = 49/84 (58%), Gaps = 2/84 (2%)
Frame = -2
Query: 229 GANLAMDNDIQKEL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTE 286
G N M+ND+ KE+ + HGK+IAQ+ L W+Y+ V F + + L IF +LT
Sbjct: 472 GPNEVMENDMLKEIVEAHGKSIAQVSLRWLYEQGVTFVAKSYDKDIMNXNLQIFDWALTX 293
Query: 287 NDFEKISEIHQERLIKEPTKPLLN 310
D EKI +I RLI PTKP LN
Sbjct: 292 EDLEKIDQIE*NRLIPGPTKPRLN 221
>TC13155 homologue to UP|Q9SXT0 (Q9SXT0) Chalcone reductase (Fragment),
partial (76%)
Length = 392
Score = 55.8 bits (133), Expect = 9e-09
Identities = 29/56 (51%), Positives = 38/56 (67%), Gaps = 1/56 (1%)
Frame = +3
Query: 256 WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLIKEPTKPLLN 310
W+Y+ + F + + L IF SLTE+D++KISEIHQ+RLIK PTKPLLN
Sbjct: 6 WLYEQGLTFVAQSYDKERMNQNLQIFDWSLTEDDYKKISEIHQDRLIKGPTKPLLN 173
>TC9510 similar to UP|O22709 (O22709) F8A5.23 protein, partial (49%)
Length = 694
Score = 36.6 bits (83), Expect = 0.006
Identities = 46/197 (23%), Positives = 85/197 (42%), Gaps = 11/197 (5%)
Frame = +3
Query: 40 VVLEAVKQGFKHFDAAAAY----AVEQSVGAAIAEEHQH*L-IASKDGLFVTSKV----W 90
++ A +G FD A Y A E VG A+ + H+ + +A+K G+ T
Sbjct: 159 IIKYAFNKGITFFDTADVYGAAGANELLVGKALKQLHREKVQLATKFGISRTQSRDFSGM 338
Query: 91 VIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*S 150
++ P+ + ++ L+ L +EY+DL+ H + PIE G
Sbjct: 339 IVKGSPDYVRSCIEASLKRLDVEYIDLYYQH------RVDTSVPIE-------DTVGELK 479
Query: 151 SMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQV--KLRDFCKEK 208
+ E +++ K IG++ S + + I+ V Q+E L + + ++ C+E
Sbjct: 480 KLVEEEKI---KYIGLSEASPDTIRRAHAVHPITAV--QIEWSLWTRDIEEEIVPLCREL 644
Query: 209 ALYWQFGIIAFSPLRMG 225
+ GI+ +SPL G
Sbjct: 645 GI----GIVPYSPLGRG 683
>TC9978 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (36%)
Length = 813
Score = 35.8 bits (81), Expect = 0.010
Identities = 30/106 (28%), Positives = 50/106 (46%), Gaps = 4/106 (3%)
Frame = +1
Query: 200 KLRDFCKEKALYWQFGIIAFSPLRMGVSRGANLAMDNDIQKELDTHGKTIAQICL*WIYK 259
KL FCK K ++ + +SPL G+ +N+ + + + GKT AQI L W
Sbjct: 4 KLHAFCKSKGVH----LSGYSPLGKGLE--SNILKNQVLHTIAEKLGKTPAQIALRW--- 156
Query: 260 HVNFCDEEL*QGEDATKLA----IFGCSLTENDFEKISEIHQERLI 301
+ L + +AT++ +F S+ + SEI QER++
Sbjct: 157 GLQMGHSVLPKSTNATRIKENIDLFDWSIPADLLANFSEIEQERIV 294
>AV426510
Length = 278
Score = 29.3 bits (64), Expect = 0.91
Identities = 12/24 (50%), Positives = 18/24 (75%), Gaps = 1/24 (4%)
Frame = +3
Query: 188 DQVE-VDLGWQQVKLRDFCKEKAL 210
DQ E +++ W QVKLR+FC + A+
Sbjct: 81 DQEEGIEVAWNQVKLRNFCDDPAM 152
>TC18006 weakly similar to UP|Q9M338 (Q9M338) Reductase-like protein,
partial (21%)
Length = 481
Score = 28.5 bits (62), Expect = 1.6
Identities = 14/24 (58%), Positives = 16/24 (66%)
Frame = +1
Query: 279 IFGCSLTENDFEKISEIHQERLIK 302
+F S+ E F K SEI QERLIK
Sbjct: 64 VFDWSIPEGLFAKFSEIKQERLIK 135
>TC14101 homologue to UP|Q39640 (Q39640) Glycolate oxidase , complete
Length = 1680
Score = 26.2 bits (56), Expect = 7.7
Identities = 24/86 (27%), Positives = 39/86 (44%), Gaps = 2/86 (2%)
Frame = +3
Query: 169 LSIKKLDKLFCFATISPVIDQVEVDLGWQQVK-LRDFCKEKALYWQFGIIAFSPLRMGVS 227
L++ +DK S V Q++ L WQ VK L+ K L G++ R+ V
Sbjct: 855 LNLGSMDKADDSGLASYVAGQIDRTLSWQDVKWLQTITKLPILV--KGVLTAEDTRIAVQ 1028
Query: 228 RG-ANLAMDNDIQKELDTHGKTIAQI 252
G A + + N ++LD TI+ +
Sbjct: 1029SGAAGIIVSNHGARQLDYVPATISAL 1106
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.333 0.144 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,686,449
Number of Sequences: 28460
Number of extensions: 55394
Number of successful extensions: 399
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 382
length of query: 316
length of database: 4,897,600
effective HSP length: 90
effective length of query: 226
effective length of database: 2,336,200
effective search space: 527981200
effective search space used: 527981200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0215b.5


