

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.11
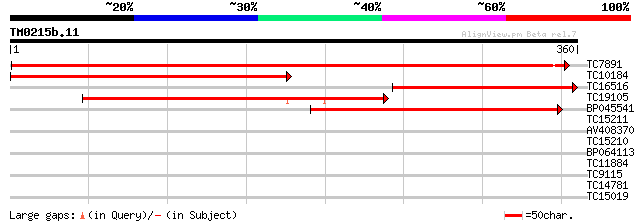
(360 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7891 similar to UP|DCAM_VICFA (Q9M4D8) S-adenosylmethionine de... 469 e-133
TC10184 similar to UP|DCAM_PHANI (Q96471) S-adenosylmethionine d... 357 2e-99
TC16516 similar to UP|Q8LKJ7 (Q8LKJ7) S-adenosylmethionine decar... 248 1e-66
TC19105 similar to UP|DCAM_HELAN (O65354) S-adenosylmethionine d... 179 6e-46
BP045541 177 3e-45
TC15211 similar to UP|Q9ZRD9 (Q9ZRD9) GMFP4 (Fragment), partial ... 30 0.82
AV408370 29 1.1
TC15210 similar to UP|Q9ZRD9 (Q9ZRD9) GMFP4 (Fragment), partial ... 28 3.1
BP064113 27 5.3
TC11884 27 5.3
TC9115 similar to PIR|T00425|T00425 photolyase/blue-light recept... 27 6.9
TC14781 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional... 26 9.1
TC15019 UP|Q9FY16 (Q9FY16) Ferredoxin-nitrite reductase precurso... 26 9.1
>TC7891 similar to UP|DCAM_VICFA (Q9M4D8) S-adenosylmethionine
decarboxylase proenzyme (AdoMetDC) (SamDC) [Contains:
S-adenosylmethionine decarboxylase alpha chain;
S-adenosylmethionine decarboxylase beta chain] ,
complete
Length = 1889
Score = 469 bits (1207), Expect = e-133
Identities = 230/356 (64%), Positives = 280/356 (78%), Gaps = 2/356 (0%)
Frame = +1
Query: 2 ALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSND 61
A+ SAIGFEGYEKRLEI+F+ G+F+DPEG GLR L++ QLDEIL PA+CTIV SL+ND
Sbjct: 541 AMAVSAIGFEGYEKRLEITFFPPGLFSDPEGKGLRALTKSQLDEILAPAECTIVSSLAND 720
Query: 62 YVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFP 121
VDSYVLSESSLFVY YK+IIKTCGTTKLLL+IP ILKLA+ +++ VKSVRYTRGSFIFP
Sbjct: 721 DVDSYVLSESSLFVYAYKIIIKTCGTTKLLLAIPPILKLAEFVSLNVKSVRYTRGSFIFP 900
Query: 122 EAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYG 181
AQ FPHR+FSEEVA+L+ YFG LGSG AY+MG D Q WH+Y+ASAE ++VY
Sbjct: 901 GAQSFPHRSFSEEVAILDGYFGKLGSGSSAYIMGGSDSEQKWHVYSASAESVRPCDSVYT 1080
Query: 182 LEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGS 241
LEMCMTGLD+E A VF+KD + SAA+MT NSGI ILP S ICDF+F+PCGYSMN +EG
Sbjct: 1081LEMCMTGLDREKAQVFYKDQSGSAAMMTVNSGIRNILPNSQICDFDFEPCGYSMNSVEGD 1260
Query: 242 AISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLD 301
A+STIHVTPEDGFSYASFE VGY+ + + L ELV RVLACF P+EFS+A+H D +
Sbjct: 1261AVSTIHVTPEDGFSYASFETVGYNLKAVDLKELVVRVLACFEPSEFSIAVHADNASRSFE 1440
Query: 302 K-FPLDIEGYYCDKRGTEELGAGGAVMFHSFVR-VDGCASPKSILKCCWSEDENDE 355
+ LD++GY C++R E LG GG+V++ FV+ C SP+S LK CW +D +E
Sbjct: 1441QGCVLDLKGYGCEERSLEGLGMGGSVVYPKFVKTASECGSPRSTLK-CWKDDSEEE 1605
>TC10184 similar to UP|DCAM_PHANI (Q96471) S-adenosylmethionine
decarboxylase proenzyme (AdoMetDC) (SamDC) [Contains:
S-adenosylmethionine decarboxylase alpha chain;
S-adenosylmethionine decarboxylase beta chain] , partial
(47%)
Length = 991
Score = 357 bits (915), Expect = 2e-99
Identities = 179/179 (100%), Positives = 179/179 (100%)
Frame = +1
Query: 1 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 60
MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN
Sbjct: 454 MALTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSN 633
Query: 61 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 120
DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF
Sbjct: 634 DYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIF 813
Query: 121 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAV 179
PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAV
Sbjct: 814 PEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAV 990
>TC16516 similar to UP|Q8LKJ7 (Q8LKJ7) S-adenosylmethionine decarboxylase,
partial (26%)
Length = 602
Score = 248 bits (633), Expect = 1e-66
Identities = 117/117 (100%), Positives = 117/117 (100%)
Frame = +1
Query: 244 STIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKF 303
STIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKF
Sbjct: 1 STIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKF 180
Query: 304 PLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDEEVKGI 360
PLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDEEVKGI
Sbjct: 181 PLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSEDENDEEVKGI 351
>TC19105 similar to UP|DCAM_HELAN (O65354) S-adenosylmethionine
decarboxylase proenzyme (AdoMetDC) (SamDC) [Contains:
S-adenosylmethionine decarboxylase alpha chain;
S-adenosylmethionine decarboxylase beta chain] , partial
(26%)
Length = 622
Score = 179 bits (454), Expect = 6e-46
Identities = 100/203 (49%), Positives = 126/203 (61%), Gaps = 9/203 (4%)
Frame = +1
Query: 47 LNPAQCTIVGSLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSI-PVILKLADSLN 105
L QCT+V ++ N Y D+YVLSESSLFVYP K+IIKTCGTT+LL SI P+I L
Sbjct: 1 LEAVQCTVVSAVGNSYFDAYVLSESSLFVYPTKIIIKTCGTTQLLKSITPLIFYAHTHLG 180
Query: 106 IAVKSVRYTRGSFIFPEAQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHI 165
+ + S RYTRGSFIFPE+QPFPH +F EEV L N S +A +M S WH+
Sbjct: 181 LTLCSCRYTRGSFIFPESQPFPHTSFEEEVTYLENTIPENLSLRKASIMPSKLSSHSWHV 360
Query: 166 YAASAEPKGS--SEAVYGLEMCMTGLDKESASVFF------KDNTSSAALMTTNSGINKI 217
+ A+ +P S Y +E+CMT LD A FF K S+ MT +GI++I
Sbjct: 361 FTATNDPPLSHYQNHPYTMEICMTDLDPILARKFFRRSGDGKSGDSAGKDMTELTGIDEI 540
Query: 218 LPQSDICDFEFDPCGYSMNGIEG 240
+ ICDF FDPCGYSMNG++G
Sbjct: 541 NRNALICDFAFDPCGYSMNGMDG 609
>BP045541
Length = 572
Score = 177 bits (448), Expect = 3e-45
Identities = 86/161 (53%), Positives = 112/161 (69%), Gaps = 1/161 (0%)
Frame = -1
Query: 192 ESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSAISTIHVTPE 251
E AS+F K ++ +MT NSGI KILP S I DF F+PCGYSMN +EG A+STIHVTPE
Sbjct: 572 EKASIFNKQQFGTSTMMTVNSGIRKILPDSQISDFAFEPCGYSMNSVEGGAVSTIHVTPE 393
Query: 252 DGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDKFPL-DIEGY 310
DGFSYASFE GY+ E ++L ++VERVLACF P EF VA+H+D+ K L D++GY
Sbjct: 392 DGFSYASFETCGYNLELVNLNQMVERVLACFPPEEFMVAVHVDVDCHSFVKDCLVDVKGY 213
Query: 311 YCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCCWSED 351
+R ++LG GG+V+FH F + C SP S CW+++
Sbjct: 212 GLQERCYQDLGMGGSVVFHKFKKTTDCGSPVSSPLECWNKE 90
>TC15211 similar to UP|Q9ZRD9 (Q9ZRD9) GMFP4 (Fragment), partial (94%)
Length = 531
Score = 29.6 bits (65), Expect = 0.82
Identities = 18/64 (28%), Positives = 31/64 (48%)
Frame = +2
Query: 50 AQCTIVGSLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVK 109
++CTI+ SLS Y + + LF++ + ++ C LLLS L D++ +A
Sbjct: 308 SRCTIM*SLSGCYSKPWPIDFLLLFLWSMPLRLEICWKKSLLLSFCTTRILLDNMTLATS 487
Query: 110 SVRY 113
Y
Sbjct: 488 CSFY 499
>AV408370
Length = 417
Score = 29.3 bits (64), Expect = 1.1
Identities = 11/14 (78%), Positives = 13/14 (92%)
Frame = +2
Query: 9 GFEGYEKRLEISFY 22
GFEGYEKRLE+ F+
Sbjct: 305 GFEGYEKRLELHFF 346
>TC15210 similar to UP|Q9ZRD9 (Q9ZRD9) GMFP4 (Fragment), partial (54%)
Length = 523
Score = 27.7 bits (60), Expect = 3.1
Identities = 17/64 (26%), Positives = 30/64 (46%)
Frame = +1
Query: 50 AQCTIVGSLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVK 109
++CTI+ SL Y + + LF++ + ++ C LLLS L D++ +A
Sbjct: 169 SRCTIM*SLCGCYSKPWQIDFLLLFLWSMPLRLEICWKKSLLLSFCTTRILLDNMTLATS 348
Query: 110 SVRY 113
Y
Sbjct: 349 CSFY 360
>BP064113
Length = 480
Score = 26.9 bits (58), Expect = 5.3
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = -3
Query: 128 HRNFSEEVAVLNNYFGNLGSGGQAYV 153
H + EV L +YF +L +GGQ+ V
Sbjct: 304 HGDMQPEVEALTHYFSSLDTGGQSMV 227
>TC11884
Length = 557
Score = 26.9 bits (58), Expect = 5.3
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +1
Query: 128 HRNFSEEVAVLNNYFGNLGSGGQAYV 153
H + EV L +YF +L +GGQ+ V
Sbjct: 448 HGDMQPEVEALTHYFSSLDTGGQSMV 525
>TC9115 similar to PIR|T00425|T00425 photolyase/blue-light receptor (PHR2)
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (55%)
Length = 1018
Score = 26.6 bits (57), Expect = 6.9
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Frame = -1
Query: 318 EELGAGGAVMFHS--FVRVDGCASPKSILKCCWSEDEN 353
E+LG G V+ S +VRV+ ++L CC E+++
Sbjct: 328 EDLGGEGEVVVESERWVRVEAWVGTLTLLGCCCGEEDD 215
>TC14781 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase (Aspartate kinase) , partial (40%)
Length = 1084
Score = 26.2 bits (56), Expect = 9.1
Identities = 11/25 (44%), Positives = 15/25 (60%)
Frame = +3
Query: 119 IFPEAQPFPHRNFSEEVAVLNNYFG 143
I+P+A+P PH F E + YFG
Sbjct: 6 IYPQAEPVPHLTFDEAAELA--YFG 74
>TC15019 UP|Q9FY16 (Q9FY16) Ferredoxin-nitrite reductase precursor ,
complete
Length = 2098
Score = 26.2 bits (56), Expect = 9.1
Identities = 13/35 (37%), Positives = 19/35 (54%)
Frame = +2
Query: 147 SGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYG 181
SGG+ +V+G ++Q WH A E + E YG
Sbjct: 143 SGGEKWVLGFEGRAQGWH*SAGKGE---AGERAYG 238
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,183,241
Number of Sequences: 28460
Number of extensions: 82178
Number of successful extensions: 373
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 365
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 367
length of query: 360
length of database: 4,897,600
effective HSP length: 91
effective length of query: 269
effective length of database: 2,307,740
effective search space: 620782060
effective search space used: 620782060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0215b.11


