

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215a.1
(623 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
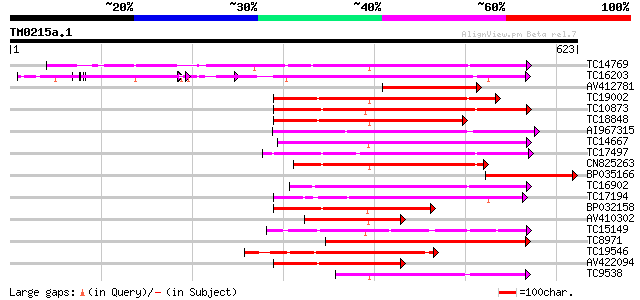
Score E
Sequences producing significant alignments: (bits) Value
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 260 5e-70
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 253 5e-68
AV412781 223 9e-59
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 215 1e-56
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 214 4e-56
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 206 1e-53
AI967315 200 5e-52
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 191 2e-49
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 184 3e-47
CN825263 182 1e-46
BP035166 179 2e-45
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 178 3e-45
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 168 2e-42
BP032158 168 3e-42
AV410302 165 2e-41
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 163 7e-41
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 159 2e-39
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 157 5e-39
AV422094 148 2e-36
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 145 2e-35
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 260 bits (664), Expect = 5e-70
Identities = 179/555 (32%), Positives = 272/555 (48%), Gaps = 22/555 (3%)
Frame = +3
Query: 41 KELLHDPRG--VLDNWDGDAVDPCSWAMVTCSSDNLVISLGTPSQNLSGTLSATIGNLTN 98
+ELL G L++W GD W + C N S+ I L
Sbjct: 1482 EELLLQNSGNRALESWSGDPCILLPWKGIACDGSNG---------------SSVITKLD- 1613
Query: 99 LQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLV 158
L ++N+ G IPS++ ++ L+TL++S+NSF G V PS L + L+ N L+
Sbjct: 1614 -----LSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSV-PSFPLSSLLISVDLSYNDLM 1775
Query: 159 GECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSILGNPLVCATGKEPNCHGITLMPM 218
G+ PES+ + L L N P ++P M
Sbjct: 1776 GKLPESIVKLPHLKSLYFGCNEHMSP---------------------EDP-------ANM 1871
Query: 219 SMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILLGFGAFLWWRHKH----------- 267
+ +L NT+ GK I G LI L FG R++
Sbjct: 1872 NSSLINTDYGRCKGKESRFGQVIVIGAITCGSLLITLAFGVLFVCRYRQKLIPWEGFAGK 2051
Query: 268 ----NQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVYKGVLS 323
F + + + +++ F +++AT + K ++G+GGFG+VY+G L+
Sbjct: 2052 KYPMETNIIFSLPSKDDFFIKSVSIQAFTLEYIEVATERY--KTLIGEGGFGSVYRGTLN 2225
Query: 324 DGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNG 383
DG VAVK ++ + G +F E+ ++S H NL+ L G+C +++LVYP+MSNG
Sbjct: 2226 DGQEVAVK-VRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNG 2402
Query: 384 SVALRLKGKP----VLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCE 439
S+ RL G+P +LDW TR IALGAARGL YLH +IHRD+K++NILLD
Sbjct: 2403 SLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMC 2582
Query: 440 AVVGDFGLAKLLDHQ-DSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITG 498
A V DFG +K + DS+V+ VRGT G++ PEY T Q SEK+DVF FG++LLE+++G
Sbjct: 2583 AKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSG 2762
Query: 499 QRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYL 558
+ L K + ++++W K++ +VD +K Y + +V+VAL C +
Sbjct: 2763 REPLNI-KRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPF 2939
Query: 559 PGHRPKMSEVVRMLE 573
+RP M +VR LE
Sbjct: 2940 STYRPSMVAIVRELE 2984
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 253 bits (647), Expect = 5e-68
Identities = 177/516 (34%), Positives = 258/516 (49%), Gaps = 20/516 (3%)
Frame = +2
Query: 77 SLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGE 136
SL + G + + + L V + NN++GPIP+ + L +DLS N+ +GE
Sbjct: 1568 SLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGE 1747
Query: 137 VPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR-----ILAK 191
VP + +L L L L+ N + G P+ + MT L+ LDLS NN +G VP +
Sbjct: 1748 VPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNY 1927
Query: 192 SFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKM-AIAFGLSLGCL 250
+ GNP +C + +C + D+L + KT ++ AI G++L
Sbjct: 1928 DKTFAGNPNLCFPHR-ASCPSVLY-----------DSLRKTRAKTARVRAIVIGIALATA 2071
Query: 251 CLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHN----FSNK 306
L++ + R H QA+ K F+ L+I + +
Sbjct: 2072 VLLVAVTVHVVRKRRLHRAQAW----------------KLTAFQRLEIKAEDVVECLKEE 2203
Query: 307 NILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGF 366
NI+GKGG G VY+G + +GT VA+KRL + + F+ E+E + HRN+++L G+
Sbjct: 2204 NIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGY 2383
Query: 367 CMTPTERLLVYPYMSNGSVALRLKGKP--VLDWGTRKHIALGAARGLLYLHEQCDPKIIH 424
LL+Y YM NGS+ L G L W R IA+ AARGL Y+H C P IIH
Sbjct: 2384 VSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIH 2563
Query: 425 RDVKAANILLDDYCEAVVGDFGLAKLL-DHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKT 483
RDVK+ NILLD EA V DFGLAK L D S +++ G+ G+IAPEY T + EK+
Sbjct: 2564 RDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKS 2743
Query: 484 DVFGFGILLLELITGQRAL-EFGKAANQKGAMLDWVKKIHLE------KKLELLVDKDLK 536
DV+ FG++LLELI G++ + EFG + ++ WV K E L L V
Sbjct: 2744 DVYSFGVVLLELIIGRKPVGEFGDGVD----IVGWVNKTMSELSQPSDTALVLAVVDPRL 2911
Query: 537 SNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRML 572
S Y + M +A++C + + RP M EVV ML
Sbjct: 2912 SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
Score = 82.8 bits (203), Expect = 2e-16
Identities = 63/213 (29%), Positives = 97/213 (44%), Gaps = 30/213 (14%)
Frame = +2
Query: 9 VLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRG---VLDNW--DGDAVDPCS 63
VLCF L+W + + S ++ AL+ +KE + + L++W CS
Sbjct: 137 VLCFT--LIWFRWTVVYSSFS---DLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCS 301
Query: 64 WAMVTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKL 123
++ VTC + V++L L G L IG L L+ + + NN++ +PS L L L
Sbjct: 302 FSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSL 481
Query: 124 QTLDLSNNSFSGE-------------------------VPPSLGHLRSLQYLRLNNNSLV 158
+ L++S+N FSG+ +P + L L+YL L N
Sbjct: 482 KVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFS 661
Query: 159 GECPESLANMTQLSFLDLSYNNLSGPVPRILAK 191
G PES + L FL L+ N+L+G VP LAK
Sbjct: 662 GTIPESYSEFQSLEFLGLNANSLTGRVPESLAK 760
Score = 79.0 bits (193), Expect = 2e-15
Identities = 46/122 (37%), Positives = 72/122 (58%), Gaps = 1/122 (0%)
Frame = +2
Query: 78 LGTPSQNLSGTLSATIGNLTNLQTVLL-QNNNISGPIPSALGKLPKLQTLDLSNNSFSGE 136
LG + +L+G + ++ L L+ + L +N G IP A G + L+ L+++N + +GE
Sbjct: 707 LGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGE 886
Query: 137 VPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSIL 196
+PPSLG+L L L + N+L G P L++M L LDLS N+L+G +P +SFS L
Sbjct: 887 IPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIP----ESFSKL 1054
Query: 197 GN 198
N
Sbjct: 1055KN 1060
Score = 73.9 bits (180), Expect = 7e-14
Identities = 37/106 (34%), Positives = 58/106 (53%)
Frame = +2
Query: 84 NLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGH 143
NL+G + ++GNLT L ++ +Q NN++G IP L + L +LDLS N +GE+P S
Sbjct: 872 NLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSK 1051
Query: 144 LRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
L++L + N G P + ++ L L + NN S +P L
Sbjct: 1052LKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNL 1189
Score = 73.6 bits (179), Expect = 9e-14
Identities = 36/108 (33%), Positives = 59/108 (54%)
Frame = +2
Query: 82 SQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSL 141
S G + G++ NL+ + + N N++G IP +LG L KL +L + N+ +G +PP L
Sbjct: 794 SNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPEL 973
Query: 142 GHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
+ SL L L+ N L GE PES + + L+ ++ N G +P +
Sbjct: 974 SSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFI 1117
Score = 62.0 bits (149), Expect = 3e-10
Identities = 54/192 (28%), Positives = 84/192 (43%), Gaps = 9/192 (4%)
Frame = +2
Query: 70 SSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLS 129
SS ++SL +L+G + + L NL + N G +PS +G LP L+TL +
Sbjct: 974 SSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVW 1153
Query: 130 NNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
N+FS +P +LG Y + N L G P L +L ++ N GP+P+ +
Sbjct: 1154 ENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGI 1333
Query: 190 AKSFSI---------LGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMA 240
+ S+ L P+ + P+ +T+ +S N N E LPS
Sbjct: 1334 GECRSLTKIRVANNFLDGPVPPGVFQLPS---VTITELSNNRLNGE--LPS--------- 1471
Query: 241 IAFGLSLGCLCL 252
+ G SLG L L
Sbjct: 1472 VISGESLGTLTL 1507
>AV412781
Length = 329
Score = 223 bits (567), Expect = 9e-59
Identities = 109/109 (100%), Positives = 109/109 (100%)
Frame = +2
Query: 410 GLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHI 469
GLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHI
Sbjct: 2 GLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHI 181
Query: 470 APEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWV 518
APEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWV
Sbjct: 182 APEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWV 328
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 215 bits (548), Expect = 1e-56
Identities = 117/253 (46%), Positives = 163/253 (64%), Gaps = 4/253 (1%)
Frame = +1
Query: 291 FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVE 350
F +EL AT+NF+ N LG+GGFG+VY G L DG+ +AVKRLK + +++F EVE
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSN-KADMEFAVEVE 204
Query: 351 MISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKP----VLDWGTRKHIALG 406
+++ H+NLL L G+C ERL+VY YM N S+ L G+ +LDW R +IA+G
Sbjct: 205 ILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIG 384
Query: 407 AARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTV 466
+A G++YLH Q P IIHRD+KA+N+LLD +A V DFG AKL+ +HVTT V+GT+
Sbjct: 385 SAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTL 564
Query: 467 GHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKK 526
G++APEY G+++E DVF FGILLLEL +G++ LE ++ K ++ DW + KK
Sbjct: 565 GYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLE-KLSSTVKRSINDWALPLACAKK 741
Query: 527 LELLVDKDLKSNY 539
D L Y
Sbjct: 742 FTEFADPRLNGEY 780
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 214 bits (544), Expect = 4e-56
Identities = 121/289 (41%), Positives = 178/289 (60%), Gaps = 6/289 (2%)
Frame = +1
Query: 291 FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVE 350
F FREL AT NF N++G+GGFG VYKG L+ G VAVK+L G + +F EV
Sbjct: 415 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQ-EFVMEVL 591
Query: 351 MISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRL----KGKPVLDWGTRKHIALG 406
M+SL H NL++L G+C +RLLVY YM GS+ L K L+W TR +A+G
Sbjct: 592 MLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVG 771
Query: 407 AARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQD-SHVTTAVRGT 465
AARGL YLH DP +I+RD+K+ANILLD+ + DFGLAKL D +HV+T V GT
Sbjct: 772 AARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGT 951
Query: 466 VGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLE- 524
G+ APEY +G+ + K+D++ FG++LLEL+TG+RA++ + ++ ++ W + +
Sbjct: 952 YGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQN-LVSWARPYFSDR 1128
Query: 525 KKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLE 573
++ +VD L+ + L + + + +C Q P RP ++++V LE
Sbjct: 1129RRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 1275
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 206 bits (523), Expect = 1e-53
Identities = 107/217 (49%), Positives = 148/217 (67%), Gaps = 4/217 (1%)
Frame = +2
Query: 291 FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVE 350
F ++EL AT FS+ N LG+GGFG+VY G SDG +AVK+LK N+ E++F EVE
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNS-KAEMEFAVEVE 442
Query: 351 MISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKPV----LDWGTRKHIALG 406
++ H+NLL L G+C+ +RL+VY YM N S+ L G+ L+W R IA+G
Sbjct: 443 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIG 622
Query: 407 AARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTV 466
+A G+LYLH + P IIHRD+KA+N+LL+ E +V DFG AKL+ SH+TT V+GT+
Sbjct: 623 SAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTL 802
Query: 467 GHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALE 503
G++APEY G+ SE DV+ FGILLLEL+TG++ +E
Sbjct: 803 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE 913
>AI967315
Length = 1308
Score = 200 bits (509), Expect = 5e-52
Identities = 124/299 (41%), Positives = 174/299 (57%), Gaps = 5/299 (1%)
Frame = +1
Query: 289 KRFPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRL-KDGNAIGGEIQFQT 347
K F + EL AT+ FS++N++GKGG+ VYKG L G +AVKRL + E +F T
Sbjct: 106 KCFSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLT 285
Query: 348 EVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKPV--LDWGTRKHIAL 405
E+ I H N++ L G C+ LV+ + GSVA + + + LDW TR I L
Sbjct: 286 EIGTIGHVCHSNVMPLLGCCID-NGLYLVFELSTVGSVASLIHDEKMAPLDWKTRYKIVL 462
Query: 406 GAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTA-VRG 464
G ARGL YLH+ C +IIHRD+KA+NILL + E + DFGLAK L Q +H + A + G
Sbjct: 463 GTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEG 642
Query: 465 TVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLE 524
T GH+APEY G EKTDVF FG+ LLE+I+G++ ++ ++ W K I +
Sbjct: 643 TFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVD-----GSHQSLHTWAKPILSK 807
Query: 525 KKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRML-EGDGLAERWE 582
++E LVD L+ YD + + A LC + RP MSEV+ ++ EG+ ERW+
Sbjct: 808 WEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKERWK 984
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 191 bits (486), Expect = 2e-49
Identities = 108/289 (37%), Positives = 169/289 (58%), Gaps = 10/289 (3%)
Frame = +2
Query: 295 ELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISL 354
EL+ T NF +K ++G+G +G VY L+DG VAVK+L + +F T+V M+S
Sbjct: 335 ELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSR 514
Query: 355 AVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGK---------PVLDWGTRKHIAL 405
+ N ++L+G+C+ R+L Y + + GS+ L G+ P LDW R IA+
Sbjct: 515 LKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAV 694
Query: 406 GAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHV-TTAVRG 464
AARGL YLHE+ P IIHRD++++N+L+ + +A + DF L+ + + +T V G
Sbjct: 695 DAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLG 874
Query: 465 TVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLE 524
T G+ APEY TGQ ++K+DV+ FG++LLEL+TG++ ++ Q+ +++ W E
Sbjct: 875 TFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQ-SLVTWATPRLSE 1051
Query: 525 KKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLE 573
K++ VD LK Y + ++ VA LC QY RP MS VV+ L+
Sbjct: 1052DKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQ 1198
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 184 bits (468), Expect = 3e-47
Identities = 115/299 (38%), Positives = 171/299 (56%), Gaps = 1/299 (0%)
Frame = +1
Query: 278 RHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGN 337
R E++ L + F F + AT++FS N LG+GGFG VYKG+L++G +AVKRL + +
Sbjct: 1603 RGDEDIDLATI--FDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTS 1776
Query: 338 AIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKPVLDW 397
G E +F+ E+++I+ HRNL+KL+G + E M + L ++DW
Sbjct: 1777 GQGME-EFKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMK---ILLDSTRSKLVDW 1944
Query: 398 GTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKL-LDHQDS 456
R I G ARGLLYLH+ +IIHRD+K +NILLDD + DFGLA++ + Q
Sbjct: 1945 NKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVE 2124
Query: 457 HVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLD 516
T V GT G++ PEY G S K+DVF FG+++LE+I+G++ F + +L
Sbjct: 2125 ARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLN-LLS 2301
Query: 517 WVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGD 575
++ +E++ LVD+ L E+ + VALLC Q P +RP M +V ML G+
Sbjct: 2302 HAWRLWIEERPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGE 2478
>CN825263
Length = 663
Score = 182 bits (463), Expect = 1e-46
Identities = 102/219 (46%), Positives = 137/219 (61%), Gaps = 5/219 (2%)
Frame = +1
Query: 313 GFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTE 372
GFG VYKG+L+DG VAVK LK + GG +F EVEM+S HRNL+KL G C+
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGR-EFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 373 RLLVYPYMSNGSVALRLKGKPV----LDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVK 428
R L+Y + NGSV L G LDW R IALGAARGL YLHE +P +IHRD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 429 AANILLDDYCEAVVGDFGLAK-LLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFG 487
++NILL+ V DFGLA+ LD + H++T V GT G++APEY TG K+DV+
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 488 FGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKK 526
+G++LLEL+TG + ++ + Q+ ++ W + I K+
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQEN-LVTWARPILTSKE 651
>BP035166
Length = 461
Score = 179 bits (453), Expect = 2e-45
Identities = 85/100 (85%), Positives = 95/100 (95%)
Frame = -1
Query: 524 EKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEA 583
EK+LELL+DKDLK+NYD+IELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGL ++WEA
Sbjct: 461 EKQLELLIDKDLKNNYDRIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLVDKWEA 282
Query: 584 SQRADTSKCRPHESSSSDRYSDLTDDSLLLVQAMELSGPR 623
SQ ADT+KC+PHE S+S+RYSDLTDDS LVQAMELSGPR
Sbjct: 281 SQNADTTKCKPHELSASNRYSDLTDDSSFLVQAMELSGPR 162
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 178 bits (451), Expect = 3e-45
Identities = 107/271 (39%), Positives = 156/271 (57%), Gaps = 5/271 (1%)
Frame = +3
Query: 308 ILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIG--GEIQFQTEVEMISLAVHRNLLKLYG 365
+LG GGFG VY G + GT VA+KR GN + G +FQTE+EM+S HR+L+ L G
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKR---GNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIG 182
Query: 366 FCMTPTERLLVYPYMSNGSVALRL--KGKPVLDWGTRKHIALGAARGLLYLHEQCDPKII 423
+C TE +LVY +M+ G++ L KP L W R I +GAARGL YLH II
Sbjct: 183 YCEENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTII 362
Query: 424 HRDVKAANILLDDYCEAVVGDFGLAKLLDHQD-SHVTTAVRGTVGHIAPEYLSTGQSSEK 482
HRDVK NILLD+ A V DFGL+K D +HV+T V+G+ G++ PEY Q ++K
Sbjct: 363 HRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDK 542
Query: 483 TDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKSNYDQI 542
+DV+ FG++L E++ + AL A ++ ++ +W + + L+ ++D LK
Sbjct: 543 SDVYSFGVVLFEILCARPALN-PSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIAPE 719
Query: 543 ELEEMVQVALLCTQYLPGHRPKMSEVVRMLE 573
++ + A+ C RP M +V+ LE
Sbjct: 720 CFKKFAETAMKCVSDQGIERPSMGDVLWNLE 812
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 168 bits (426), Expect = 2e-42
Identities = 109/286 (38%), Positives = 162/286 (56%), Gaps = 7/286 (2%)
Frame = +1
Query: 291 FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVE 350
F ++EL AT+NFS N +G+GGFG VY L G A+K++ + +F E++
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMD----VQASTEFLCELK 1215
Query: 351 MISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRL--KGKPVLDWGTRKHIALGAA 408
+++ H NL++L G+C+ + LVY ++ NG++ L GK L W +R IAL AA
Sbjct: 1216 VLTHVHHLNLVRLIGYCVEGS-LFLVYEHIDNGNLGQYLHGSGKEPLPWSSRVQIALDAA 1392
Query: 409 RGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGH 468
RGL Y+HE P IHRDVK+ANIL+D V DFGL KL++ +S + T + GT G+
Sbjct: 1393 RGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFGY 1572
Query: 469 IAPEYLSTGQSSEKTDVFGFGILLLELITGQRA-LEFGK-AANQKGAMLDWVKKIHLE-- 524
+ PEY G S K DV+ FG++L ELI+ + A L+ G+ A KG + + + ++
Sbjct: 1573 MPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEALNKSDP 1752
Query: 525 -KKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVV 569
L LVD L NY + ++ Q+ CT+ P RP M +V
Sbjct: 1753 CDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLV 1890
>BP032158
Length = 555
Score = 168 bits (425), Expect = 3e-42
Identities = 87/181 (48%), Positives = 121/181 (66%), Gaps = 4/181 (2%)
Frame = +2
Query: 291 FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVE 350
F R+++ AT+NF N +G+GGFG VYKGVLS+G ++AVK+L + G +F E+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNR-EFINEIG 190
Query: 351 MISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKG----KPVLDWGTRKHIALG 406
MIS H NL+KLYG C+ + LLVY YM N S+A L G K L+W TR I +G
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG 370
Query: 407 AARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTV 466
A+GL YLHE+ KI+HRD+KA N+LLD +A + DFGLAKL + +++H++T + GT+
Sbjct: 371 IAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIAGTI 550
Query: 467 G 467
G
Sbjct: 551 G 553
>AV410302
Length = 349
Score = 165 bits (418), Expect = 2e-41
Identities = 83/115 (72%), Positives = 96/115 (83%), Gaps = 4/115 (3%)
Frame = +3
Query: 325 GTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGS 384
G+L+AVKRLK+ GE+QFQTEVEMIS+AVHRNLL+L GFC+TPTERLLVYP+M NGS
Sbjct: 3 GSLIAVKRLKEERTQSGELQFQTEVEMISMAVHRNLLRLRGFCITPTERLLVYPFMVNGS 182
Query: 385 VALRLKGK----PVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLD 435
+A L+G+ P LDW RK IALG+ARGL YLH+ CDPKIIHRDVKAANILLD
Sbjct: 183 LASCLRGRPESQPPLDWLMRKRIALGSARGLSYLHDYCDPKIIHRDVKAANILLD 347
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 163 bits (413), Expect = 7e-41
Identities = 114/300 (38%), Positives = 164/300 (54%), Gaps = 9/300 (3%)
Frame = +3
Query: 283 VYLGNLKR-FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGG 341
V+ GN R F +L A+ +LGKG FG YK VL G +VAVKRLKD
Sbjct: 228 VFFGNSARGFDLEDLLRAS-----AEVLGKGTFGTAYKAVLETGLVVAVKRLKDVTI--S 386
Query: 342 EIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRL-----KGKPVLD 396
E +F+ ++E + H++L+ L + + E+LLVY YM GS++ L G+ L+
Sbjct: 387 EKEFKDKIETVGAMDHQSLVPLRAYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLN 566
Query: 397 WGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDS 456
W R IALGAARG+ YLH Q P + H ++KA+NILL EA V DFGLA L+ +
Sbjct: 567 WEIRSGIALGAARGIEYLHSQ-GPNVSHGNIKASNILLTKSYEAKVSDFGLAHLVGPSST 743
Query: 457 HVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLD 516
A G+ APE + S+K DV+ FG+LLLEL+TG+ N++G L
Sbjct: 744 PNRVA-----GYRAPEVTDPRKVSQKADVYSFGVLLLELLTGKAPTH--ALLNEEGVDLP 902
Query: 517 -WVKKIHLEKKLELLVDKDLKSNYDQIELE--EMVQVALLCTQYLPGHRPKMSEVVRMLE 573
WV+ + E+ + D +L Y +E E +++Q+A+ C P RP ++EV R +E
Sbjct: 903 RWVQSLVREEWTSEVFDLEL-LRYQNVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSIE 1079
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 159 bits (401), Expect = 2e-39
Identities = 90/229 (39%), Positives = 139/229 (60%), Gaps = 4/229 (1%)
Frame = +3
Query: 348 EVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGK--PVLDWGTRKHIAL 405
EVE+++ HRNL+KL GF ER+L+ Y+ NG++ L G +LD+ R IA+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKILDFNQRLEIAI 185
Query: 406 GAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKL--LDHQDSHVTTAVR 463
A GL YLH + +IIHRDVK++NILL + A V DFG A+L ++ +H++T V+
Sbjct: 186 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTKVK 365
Query: 464 GTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHL 523
GTVG++ PEY+ T Q + K+DV+ FGILLLE++TG+R +E KAA+++ L W + +
Sbjct: 366 GTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADER-VTLRWAFRKYN 542
Query: 524 EKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRML 572
E + L+D ++ + L +M+ ++ C + RP M V L
Sbjct: 543 EGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQL 689
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 157 bits (397), Expect = 5e-39
Identities = 91/216 (42%), Positives = 130/216 (60%), Gaps = 3/216 (1%)
Frame = +1
Query: 259 AFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVY 318
AF WW H++N Q + ++ ++++Q AT NF+ ILG+G FG VY
Sbjct: 136 AFSWWSHQNNDQFASP-----------SGIPKYSYKDIQKATQNFTT--ILGQGSFGTVY 276
Query: 319 KGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYP 378
K + G +VAVK L N+ GE +FQTEV ++ HRNL+ L GFC+ +R+LVY
Sbjct: 277 KATMPTGEVVAVKVLAP-NSKQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQ 453
Query: 379 YMSNGSVALRLKGKPV-LDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDY 437
+MSNGS+A L G+ L W R IA+ + G+ YLHE P +IHRD+K+ANILLDD
Sbjct: 454 FMSNGSLANLLYGEEKELSWDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDS 633
Query: 438 CEAVVGDFGLAK--LLDHQDSHVTTAVRGTVGHIAP 471
A+V DFGL+K + D ++S ++GT G++ P
Sbjct: 634 MRAMVADFGLSKEEIFDGRNS----GLKGTYGYMDP 729
>AV422094
Length = 462
Score = 148 bits (374), Expect = 2e-36
Identities = 79/146 (54%), Positives = 105/146 (71%), Gaps = 1/146 (0%)
Frame = +2
Query: 291 FPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVE 350
F + EL+ AT++F+ N LG+GGFG VYKG+L+DGT++AVK+L G+ G+ QF E+
Sbjct: 14 FSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTVIAVKQLSLGSH-QGKSQFIAEIA 190
Query: 351 MISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKPV-LDWGTRKHIALGAAR 409
IS HRNL+KLYG C+ ++RLLVY Y+ N S+ L GK + L+W TR I LG AR
Sbjct: 191 TISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKALSLNWSTRYDICLGVAR 370
Query: 410 GLLYLHEQCDPKIIHRDVKAANILLD 435
GL YLHE+ +I+HRDVKA+NILLD
Sbjct: 371 GLTYLHEESRLRIVHRDVKASNILLD 448
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 145 bits (366), Expect = 2e-35
Identities = 88/229 (38%), Positives = 128/229 (55%), Gaps = 15/229 (6%)
Frame = +2
Query: 359 NLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKP-------------VLDWGTRKHIAL 405
N+++L LLVY Y+ N S+ L KP VLDW R IA+
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 406 GAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAK-LLDHQDSHVTTAVRG 464
GAA+GL Y+H C P I+HRDVK +NILLD A V DFGLA+ L+ + ++ + V G
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIG 364
Query: 465 TVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWV-KKIHL 523
T G+IAPEY+ T + SEK DV+ FG++LLEL TG+ A +Q ++ +W + I +
Sbjct: 365 TFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKEA----NYGDQHSSLAEWAWRHILI 532
Query: 524 EKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRML 572
+ L+ KD+ E+ + ++ ++CT LP RP M EV+ +L
Sbjct: 533 GSNVXDLLXKDVMEASYIDEMCSVFKLGVMCTATLPATRPSMKEVLPIL 679
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,056,057
Number of Sequences: 28460
Number of extensions: 157165
Number of successful extensions: 1602
Number of sequences better than 10.0: 406
Number of HSP's better than 10.0 without gapping: 1159
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1296
length of query: 623
length of database: 4,897,600
effective HSP length: 96
effective length of query: 527
effective length of database: 2,165,440
effective search space: 1141186880
effective search space used: 1141186880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0215a.1


