

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0212.10
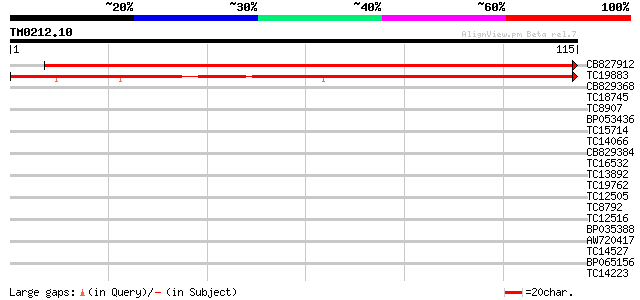
(115 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB827912 219 4e-59
TC19883 similar to AAR24691 (AAR24691) At2g18210, partial (35%) 122 6e-30
CB829368 28 0.16
TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier... 27 0.47
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 26 1.1
BP053436 26 1.1
TC15714 weakly similar to UP|Q9LU96 (Q9LU96) Genomic DNA, chromo... 25 1.4
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 25 1.4
CB829384 25 1.8
TC16532 similar to UP|Q9LSV4 (Q9LSV4) Gb|AAB61039.1, partial (9%) 25 1.8
TC13892 25 1.8
TC19762 similar to UP|Q9FKP4 (Q9FKP4) Similarity to kinesin heav... 25 2.4
TC12505 similar to UP|BAD00698 (BAD00698) Sericin 1A, partial (5%) 25 2.4
TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosid... 24 3.1
TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like pr... 24 3.1
BP035388 24 4.0
AW720417 24 4.0
TC14527 similar to PIR|F86340|F86340 protein F2D10.34 [imported]... 24 4.0
BP065156 24 4.0
TC14223 similar to UP|Q8GZV0 (Q8GZV0) Obtusifoliol-14-demethylas... 23 5.2
>CB827912
Length = 348
Score = 219 bits (559), Expect = 4e-59
Identities = 108/108 (100%), Positives = 108/108 (100%)
Frame = +3
Query: 8 RRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSK 67
RRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSK
Sbjct: 3 RRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSK 182
Query: 68 STHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHYK 115
STHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHYK
Sbjct: 183 STHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHYK 326
>TC19883 similar to AAR24691 (AAR24691) At2g18210, partial (35%)
Length = 561
Score = 122 bits (307), Expect = 6e-30
Identities = 66/121 (54%), Positives = 88/121 (72%), Gaps = 6/121 (4%)
Frame = +3
Query: 1 MLRSFTTR----RYERLGKETASSA-LLQEGFKRSTSLPSRASSSARKMAHATFGNINLQ 55
M +S +TR +YE++ K++A+ E RSTS+PS SSA MA + +INL
Sbjct: 78 MFKSLSTRPGPSKYEKMDKDSAAGGGTSNEEMTRSTSVPS---SSATSMALGS-SSINLH 245
Query: 56 RNPTKKA-NNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHY 114
RNPTK+A NN + +HP+FS +FRRKKK TA+PEFTRYLEY+KEGGMWD+NSN+PVI++
Sbjct: 246 RNPTKRATNNPKEKSHPIFSLFEFRRKKKATAKPEFTRYLEYMKEGGMWDVNSNQPVIYF 425
Query: 115 K 115
K
Sbjct: 426 K 428
>CB829368
Length = 541
Score = 28.5 bits (62), Expect = 0.16
Identities = 17/51 (33%), Positives = 25/51 (48%), Gaps = 4/51 (7%)
Frame = -2
Query: 52 INLQRNPTKKANNSSKSTHPLFSFLDFRRKKKTTAR----PEFTRYLEYLK 98
+ +QRN KK + S T+P F FL R + + R P T + E +K
Sbjct: 399 LRMQRNEAKKFSLSESPTNPTFLFLIKPRNRNGSIRGKSQPRGTHFQELMK 247
>TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier protein,
fibroblast isoform (ADP/ATP translocase 2) (Adenine
nucleotide translocator 2) (ANT 2), partial (9%)
Length = 834
Score = 26.9 bits (58), Expect = 0.47
Identities = 15/39 (38%), Positives = 24/39 (61%)
Frame = -3
Query: 18 ASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
A+SAL F S ++P+RAS +A ++ H+ +I L R
Sbjct: 229 ATSALTGLCFPPSITMPTRASMTATQLTHSINQSIMLYR 113
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 25.8 bits (55), Expect = 1.1
Identities = 12/35 (34%), Positives = 23/35 (65%)
Frame = +1
Query: 4 SFTTRRYERLGKETASSALLQEGFKRSTSLPSRAS 38
S +TR KET+ + ++E ++++ +LPSR+S
Sbjct: 334 STSTRLARS*AKETSPRSTMEETWRQTRALPSRSS 438
>BP053436
Length = 463
Score = 25.8 bits (55), Expect = 1.1
Identities = 8/17 (47%), Positives = 12/17 (70%)
Frame = -1
Query: 99 EGGMWDLNSNKPVIHYK 115
E G+W L N+P+ HY+
Sbjct: 220 ESGLWSLKCNQPLPHYE 170
>TC15714 weakly similar to UP|Q9LU96 (Q9LU96) Genomic DNA, chromosome 3,
P1 clone: MPE11, partial (26%)
Length = 1157
Score = 25.4 bits (54), Expect = 1.4
Identities = 12/30 (40%), Positives = 20/30 (66%)
Frame = +1
Query: 53 NLQRNPTKKANNSSKSTHPLFSFLDFRRKK 82
N ++NP + +++SSKS L FL F++ K
Sbjct: 16 NPKQNPHQSSSSSSKSESKLRIFLQFQKIK 105
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 25.4 bits (54), Expect = 1.4
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Frame = -2
Query: 30 STSLPSRASSSARKMAHATFGN-INLQRNPTKKANNSSKSTHPLFSFLD 77
S S P +SSS R M + N I+ + N + N+S++ PL SFLD
Sbjct: 535 SASSPIPSSSSKRDMGSSQRSNAISGRENKDIRTGNNSRAL-PLQSFLD 392
>CB829384
Length = 392
Score = 25.0 bits (53), Expect = 1.8
Identities = 15/36 (41%), Positives = 21/36 (57%), Gaps = 1/36 (2%)
Frame = -1
Query: 54 LQRNPTKKANNSSKSTHPLFS-FLDFRRKKKTTARP 88
LQR + KA + ++S + LFS F RR+ TA P
Sbjct: 311 LQRTKSAKARSGARSLNSLFSLFT*IRRQTARTAVP 204
>TC16532 similar to UP|Q9LSV4 (Q9LSV4) Gb|AAB61039.1, partial (9%)
Length = 471
Score = 25.0 bits (53), Expect = 1.8
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 2/57 (3%)
Frame = +3
Query: 18 ASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR--NPTKKANNSSKSTHPL 72
ASS+LL ++TS P+ S ++ H T +NL P +K ++ +S H L
Sbjct: 63 ASSSLLIHSATKTTSKPTNPSLLSQTTLHTTL-QLNLLHLLLPPRKTSSKIQSFHSL 230
>TC13892
Length = 519
Score = 25.0 bits (53), Expect = 1.8
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 8/50 (16%)
Frame = -3
Query: 71 PLFSFLD------FRRKKKTTARPEFTRYLEYLKEGGMWDLN--SNKPVI 112
P+ SFLD F + +K P+F LE + +WDL+ +PV+
Sbjct: 223 PMVSFLD*VHLLAFHQTRKK*LAPQF*LLLEEARAPQLWDLHWKHTRPVL 74
>TC19762 similar to UP|Q9FKP4 (Q9FKP4) Similarity to kinesin heavy chain,
partial (22%)
Length = 838
Score = 24.6 bits (52), Expect = 2.4
Identities = 12/48 (25%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Frame = -3
Query: 63 NNSSKSTHPLFSFLDFR-RKKKTTARPEFTRYLEYLKEGGMWDLNSNK 109
N+++ + H + + L F K+ + P F++YL + + G +D + N+
Sbjct: 194 NSNTGNKHGVHNSLWFHFLNKRENSIPNFSKYLMFSRGTGFYDSSQNQ 51
>TC12505 similar to UP|BAD00698 (BAD00698) Sericin 1A, partial (5%)
Length = 569
Score = 24.6 bits (52), Expect = 2.4
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 3/65 (4%)
Frame = +2
Query: 25 EGFKRSTSLPSRASSSARKMAHAT---FGNINLQRNPTKKANNSSKSTHPLFSFLDFRRK 81
+GF+ +T+ S +SSS+R A ++ FG + + ++SS ST S F
Sbjct: 320 DGFENATTSSSSSSSSSRSSASSSLFLFGCEEVAAAGSTWTSSSSSSTSSSSSSSPFSFT 499
Query: 82 KKTTA 86
T+A
Sbjct: 500 SSTSA 514
>TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase {Prunus
persica;} , partial (90%)
Length = 1636
Score = 24.3 bits (51), Expect = 3.1
Identities = 14/41 (34%), Positives = 19/41 (46%)
Frame = +3
Query: 35 SRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSF 75
S SS A K++HA G + + + K S TH L F
Sbjct: 981 STMSSKAVKVSHAKCGALKMGFHVDVKNEGSMDGTHTLLIF 1103
>TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like protein
(Fragment), partial (36%)
Length = 859
Score = 24.3 bits (51), Expect = 3.1
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +1
Query: 80 RKKKTTARPEFTRYLEYLKEGGMWDLNSNK 109
R KKTT +P FTR+ + G L S++
Sbjct: 139 RMKKTTRKPLFTRWKLPINRGNSTILKSSQ 228
>BP035388
Length = 557
Score = 23.9 bits (50), Expect = 4.0
Identities = 14/48 (29%), Positives = 21/48 (43%)
Frame = +3
Query: 55 QRNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGM 102
Q++P K NN+ H S + + T +PE LE + GM
Sbjct: 21 QKDPNTKNNNAPSPHHGCSSEKSEPVRSRWTPKPEQILILESIFNSGM 164
>AW720417
Length = 554
Score = 23.9 bits (50), Expect = 4.0
Identities = 11/31 (35%), Positives = 18/31 (57%)
Frame = +1
Query: 35 SRASSSARKMAHATFGNINLQRNPTKKANNS 65
S SSS K++ +T +IN + + +NNS
Sbjct: 91 STTSSSINKISRSTTSSINTINSSSSNSNNS 183
>TC14527 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(42%)
Length = 1095
Score = 23.9 bits (50), Expect = 4.0
Identities = 11/25 (44%), Positives = 18/25 (72%)
Frame = -1
Query: 30 STSLPSRASSSARKMAHATFGNINL 54
ST + S AS+SAR++AH+ + +L
Sbjct: 558 STRISSPASNSARQIAHSVVASHSL 484
>BP065156
Length = 470
Score = 23.9 bits (50), Expect = 4.0
Identities = 18/59 (30%), Positives = 25/59 (41%)
Frame = +2
Query: 36 RASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYL 94
+A + RK+ GNI Q N ++ LFS F K +P+FT YL
Sbjct: 125 KAQRADRKVILKIKGNIRTQTN--------TRGFSILFSLRVFGGNMKENYQPQFTWYL 277
>TC14223 similar to UP|Q8GZV0 (Q8GZV0) Obtusifoliol-14-demethylase, complete
Length = 1917
Score = 23.5 bits (49), Expect = 5.2
Identities = 12/40 (30%), Positives = 19/40 (47%)
Frame = +1
Query: 30 STSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKST 69
S PS A+SSA A ++ + + + +NSS T
Sbjct: 220 SADFPSSAASSASSKARSSCSEKSTRNSAASSPSNSSTGT 339
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,740,513
Number of Sequences: 28460
Number of extensions: 20039
Number of successful extensions: 147
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 146
length of query: 115
length of database: 4,897,600
effective HSP length: 91
effective length of query: 24
effective length of database: 2,307,740
effective search space: 55385760
effective search space used: 55385760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 47 (22.7 bits)
Lotus: description of TM0212.10


