

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0207.16
(596 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
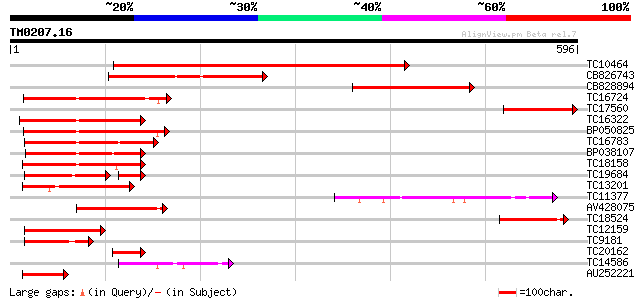
Score E
Sequences producing significant alignments: (bits) Value
TC10464 weakly similar to UP|Q8L8G0 (Q8L8G0) Nam-like protein 1,... 631 0.0
CB826743 223 7e-59
CB828894 211 2e-55
TC16724 similar to GB|BAB20599.1|12060424|AB049070 AtNAC3 {Arabi... 151 3e-37
TC17560 150 7e-37
TC16322 homologue to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial... 149 1e-36
BP050825 149 1e-36
TC16783 similar to UP|AAR88435 (AAR88435) NAC domain protein, pa... 148 2e-36
BP038107 144 3e-35
TC18158 similar to UP|Q84TD6 (Q84TD6) At3g04070, partial (34%) 144 4e-35
TC19684 similar to UP|AAR88435 (AAR88435) NAC domain protein, pa... 100 2e-30
TC13201 similar to UP|Q8LRL6 (Q8LRL6) Nam-like protein 9, partia... 126 9e-30
TC11377 UP|Q93ZN0 (Q93ZN0) AT4g35580/F8D20_90, partial (5%) 110 5e-25
AV428075 105 3e-23
TC18524 99 3e-21
TC12159 similar to UP|Q9M4U3 (Q9M4U3) NAC1, partial (21%) 96 2e-20
TC9181 similar to UP|Q9FY93 (Q9FY93) NAM-like protein (AT5g13180... 69 2e-12
TC20162 similar to UP|Q8LKN6 (Q8LKN6) Nam-like protein 18, parti... 61 6e-10
TC14586 similar to UP|Q9FY93 (Q9FY93) NAM-like protein (AT5g1318... 59 2e-09
AU252221 55 3e-08
>TC10464 weakly similar to UP|Q8L8G0 (Q8L8G0) Nam-like protein 1, partial
(12%)
Length = 935
Score = 631 bits (1628), Expect = 0.0
Identities = 311/311 (100%), Positives = 311/311 (100%)
Frame = +1
Query: 110 SGSSLIGMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDE 169
SGSSLIGMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDE
Sbjct: 1 SGSSLIGMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDE 180
Query: 170 SIEVSISGEAEQTASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEET 229
SIEVSISGEAEQTASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEET
Sbjct: 181 SIEVSISGEAEQTASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEET 360
Query: 230 TSNIITPVDRNSDRYDAQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHL 289
TSNIITPVDRNSDRYDAQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHL
Sbjct: 361 TSNIITPVDRNSDRYDAQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHL 540
Query: 290 PPVFDYQASNEPDDRSGLQYGTNETAISDLFDSLTWDQLSYEESGSQPMNFPLFNVKDSG 349
PPVFDYQASNEPDDRSGLQYGTNETAISDLFDSLTWDQLSYEESGSQPMNFPLFNVKDSG
Sbjct: 541 PPVFDYQASNEPDDRSGLQYGTNETAISDLFDSLTWDQLSYEESGSQPMNFPLFNVKDSG 720
Query: 350 SGSDSDFEITNMPSMQIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQN 409
SGSDSDFEITNMPSMQIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQN
Sbjct: 721 SGSDSDFEITNMPSMQIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQN 900
Query: 410 QNAAQMTFSSD 420
QNAAQMTFSSD
Sbjct: 901 QNAAQMTFSSD 933
>CB826743
Length = 513
Score = 223 bits (568), Expect = 7e-59
Identities = 116/167 (69%), Positives = 134/167 (79%)
Frame = +2
Query: 105 DRKIKSGSSLIGMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLF 164
DRKIKSGS+LIGMKKTLVFY GRAPKGKRT+WVMHEYRPTL+ELDGTNP QNPYV+CRLF
Sbjct: 2 DRKIKSGSTLIGMKKTLVFYAGRAPKGKRTHWVMHEYRPTLKELDGTNP*QNPYVLCRLF 181
Query: 165 KKQDESIEVSISGEAEQTASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPE 224
KK DES+E S + E EQ ASTP A +SPEE+QSD LV VSSSL TEDDKH +IPE
Sbjct: 182 KKNDESLEGS-NEEVEQIASTPTTASFSPEEIQSDP--SLVPVSSSLATEDDKHLPVIPE 352
Query: 225 TSEETTSNIITPVDRNSDRYDAQNAQPQNVKLAAEENQPLNLDMYND 271
SEE SNII PVD +SD DA +A+ Q ++ A+E+QPLN D++ D
Sbjct: 353 QSEEAISNIINPVDCHSDGGDAHDAKIQIIEPPAQEDQPLNFDIFYD 493
>CB828894
Length = 432
Score = 211 bits (538), Expect = 2e-55
Identities = 106/128 (82%), Positives = 113/128 (87%)
Frame = +3
Query: 361 MPSMQIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQNQNAAQMTFSSD 420
+ SMQ+PYPEEA+D +PLGTAPEFFS VGVPFDHS EQK NVGLFQNQN AQMTFSSD
Sbjct: 48 LQSMQVPYPEEAMDRNVPLGTAPEFFSTVGVPFDHSAVEQKGNVGLFQNQNDAQMTFSSD 227
Query: 421 VSMGQVYNVVNDYEQPRNWNVATGGDTGIIIRARQAQNELVNTNINQGSANRRIRLLKSA 480
VSMGQV NVVNDYE+PRN NV+ GDTGII RAR+ QNELVNTNINQGSANR IRLLKSA
Sbjct: 228 VSMGQVSNVVNDYEEPRNCNVSADGDTGIIRRARRPQNELVNTNINQGSANR*IRLLKSA 407
Query: 481 HGSSRVVK 488
HGSSRVVK
Sbjct: 408 HGSSRVVK 431
>TC16724 similar to GB|BAB20599.1|12060424|AB049070 AtNAC3 {Arabidopsis
thaliana;}, partial (53%)
Length = 615
Score = 151 bits (382), Expect = 3e-37
Identities = 75/162 (46%), Positives = 107/162 (65%), Gaps = 6/162 (3%)
Frame = +2
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE++ YL +K+ G + +I EID+ K++PW LP ++ + EW
Sbjct: 131 SLPPGFRFFPTDEELLVQYLCRKVAGYHFSLPIIAEIDLYKFDPWVLPGKAIFGEK--EW 304
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF P+DRKYPNG+R NR GYWKATG D+ I + +G+KK LVFY G+APKG +T
Sbjct: 305 YFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKVITTEGRKVGIKKALVFYIGKAPKGTKT 484
Query: 135 NWVMHEYRPTLQELDGTNPGQ------NPYVICRLFKKQDES 170
NW+MHEYR LD + + + +V+CR++KK+ +
Sbjct: 485 NWIMHEYR----LLDSSRKHKLGSSRLDEWVLCRIYKKKSST 598
>TC17560
Length = 467
Score = 150 bits (378), Expect = 7e-37
Identities = 76/77 (98%), Positives = 77/77 (99%)
Frame = +2
Query: 520 TLTNHVKAMPKTPKSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPCISKASSNRAMWSSV 579
TLTNHVKAMPKTPKSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPCISKASS+RAMWSSV
Sbjct: 2 TLTNHVKAMPKTPKSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPCISKASSHRAMWSSV 181
Query: 580 FVVSAFVMVSVMVFSLY 596
FVVSAFVMVSVMVFSLY
Sbjct: 182 FVVSAFVMVSVMVFSLY 232
>TC16322 homologue to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (40%)
Length = 510
Score = 149 bits (377), Expect = 1e-36
Identities = 70/132 (53%), Positives = 94/132 (71%)
Frame = +1
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
LS SLP GFRF P+DEE++ YL +K+ G+ + +I EID+ K++PW LP ++ +
Sbjct: 121 LSQLSLPPGFRFYPTDEELLVQYLCRKVAGHNFTLPIIAEIDLYKFDPWVLPSKAIFGEK 300
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPK 130
EW+FF P+DRKYPNG+R NR GYWKATG D+ I + +G+KK LVFY G+APK
Sbjct: 301 --EWYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYVGKAPK 474
Query: 131 GKRTNWVMHEYR 142
G +TNW+MHEYR
Sbjct: 475 GTKTNWIMHEYR 510
>BP050825
Length = 539
Score = 149 bits (376), Expect = 1e-36
Identities = 72/167 (43%), Positives = 106/167 (63%), Gaps = 13/167 (7%)
Frame = +2
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
+LP GFRF P+DEE++ YL++K V +I E+D+ K++PW+LP + + EW
Sbjct: 35 NLPPGFRFHPTDEELVVHYLKKKAASVPLPVAIIAEVDLYKFDPWELPAKATFGEQ--EW 208
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKI--KSGSSLIGMKKTLVFYHGRAPKGK 132
+FF P+DRKYPNG R NRA GYWKATG D+ + +G+ +G+KK LVFY G+ P+G
Sbjct: 209 YFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVLTSNGTQKVGVKKALVFYGGKPPRGI 388
Query: 133 RTNWVMHEYRPTLQELDGTNPG-----------QNPYVICRLFKKQD 168
+TNW+MHEYR + + PG + +V+CR++KK +
Sbjct: 389 KTNWIMHEYRLADNKPNNRPPGCDLGNKKNSLRLDDWVLCRIYKKSN 529
>TC16783 similar to UP|AAR88435 (AAR88435) NAC domain protein, partial (46%)
Length = 511
Score = 148 bits (374), Expect = 2e-36
Identities = 70/141 (49%), Positives = 94/141 (66%)
Frame = +1
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+D+E++ YL K V +I+E+D+ K++PW LPD+++ + EW+
Sbjct: 97 LPPGFRFHPTDDELVKHYLCAKCASQPINVPIIKELDLYKFDPWQLPDMALYGEK--EWY 270
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P+DRKYPNG+R NRA GYWKATG D+ I +L G+KK LVFY G+APKG +TN
Sbjct: 271 FFTPRDRKYPNGSRPNRAAGTGYWKATGADKPIGKPKTL-GIKKALVFYAGKAPKGVKTN 447
Query: 136 WVMHEYRPTLQELDGTNPGQN 156
W+MHEYR + T N
Sbjct: 448 WIMHEYRLANVDRSATKKNNN 510
>BP038107
Length = 469
Score = 144 bits (364), Expect = 3e-35
Identities = 69/126 (54%), Positives = 88/126 (69%)
Frame = +3
Query: 17 PTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWFF 76
P GFRF P+DEE++ YL +K V +I EID+ K++PWDLP L+ + EW+F
Sbjct: 66 PPGFRFHPTDEELVLHYLCRKCASQPIAVPIIAEIDLYKYDPWDLPGLASYGEK--EWYF 239
Query: 77 FCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTNW 136
F P+DRKYPNG+R NRA GYWKATG D+ I +G+KK LVFY G+APKG +TNW
Sbjct: 240 FSPRDRKYPNGSRPNRAAGTGYWKATGADKPI-GHPKPVGIKKALVFYAGKAPKGDKTNW 416
Query: 137 VMHEYR 142
+MHEYR
Sbjct: 417 IMHEYR 434
>TC18158 similar to UP|Q84TD6 (Q84TD6) At3g04070, partial (34%)
Length = 476
Score = 144 bits (363), Expect = 4e-35
Identities = 69/136 (50%), Positives = 91/136 (66%), Gaps = 7/136 (5%)
Frame = +3
Query: 14 NSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPE 73
++LP GFRF P+DEE+I YLR+K+ V +I E+D+ K +PW+LP ++ + E
Sbjct: 72 SNLPPGFRFHPTDEELILHYLRKKVASIPLPVSIIAEVDIYKLDPWELPGKALFGEK--E 245
Query: 74 WFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKS-------GSSLIGMKKTLVFYHG 126
W+FF P+DRKYPNG R NRA GYWKATG D+ I + IG+KK LVFY G
Sbjct: 246 WYFFSPRDRKYPNGARPNRAAASGYWKATGTDKTIVASLPGGGRAQDSIGVKKALVFYKG 425
Query: 127 RAPKGKRTNWVMHEYR 142
+ PKG +TNW+MHEYR
Sbjct: 426 KPPKGVKTNWIMHEYR 473
>TC19684 similar to UP|AAR88435 (AAR88435) NAC domain protein, partial (48%)
Length = 535
Score = 100 bits (250), Expect(2) = 2e-30
Identities = 47/91 (51%), Positives = 64/91 (69%)
Frame = +2
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+++ YL K G VI+EID+ K++PW LP++ + + + EW+
Sbjct: 110 LPPGFRFHPTDEELVNHYLCTKGAGQSFNYSVIKEIDLYKFDPWQLPEMGL--DGEKEWY 283
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDR 106
FF P+DRKYPNG+R NRA GY KATG D+
Sbjct: 284 FFSPRDRKYPNGSRPNRAAGSGYGKATGADK 376
Score = 48.9 bits (115), Expect(2) = 2e-30
Identities = 19/28 (67%), Positives = 24/28 (84%)
Frame = +3
Query: 115 IGMKKTLVFYHGRAPKGKRTNWVMHEYR 142
+G+KK LV Y G+APKG +TNW+MHEYR
Sbjct: 399 LGIKKALVLYVGKAPKGIKTNWIMHEYR 482
>TC13201 similar to UP|Q8LRL6 (Q8LRL6) Nam-like protein 9, partial (22%)
Length = 465
Score = 126 bits (317), Expect = 9e-30
Identities = 60/122 (49%), Positives = 85/122 (69%), Gaps = 4/122 (3%)
Frame = +3
Query: 14 NSLPTGFRFRPSDEEIIDFYLRQKING---NGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
++L GFRF P+DEE++ FYL++++ G +GD + ++ DV K+EPWDLP LS ++ R
Sbjct: 105 SNLSPGFRFHPTDEELVGFYLKRRVTGRLFHGDPIGIV---DVYKYEPWDLPCLSKMKTR 275
Query: 71 DPEWFFFCPQDRKYPNGN-RLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAP 129
D EW+F+ D+KY G+ R NRAT GYWK TGKDR + + +GMKKTLV++ GRA
Sbjct: 276 DLEWYFYSGLDKKYGKGSSRTNRATEKGYWKTTGKDRPVNHANRTVGMKKTLVYHFGRAS 455
Query: 130 KG 131
G
Sbjct: 456 TG 461
>TC11377 UP|Q93ZN0 (Q93ZN0) AT4g35580/F8D20_90, partial (5%)
Length = 1165
Score = 110 bits (276), Expect = 5e-25
Identities = 102/283 (36%), Positives = 136/283 (48%), Gaps = 49/283 (17%)
Frame = +1
Query: 342 LFNVKDSGSGSDSDFEITNMPSMQI-------------------PYPEEAIDMK--IPLG 380
L N+KD+GS SDSD E+ N+ I P P + +K + L
Sbjct: 43 LVNLKDNGSCSDSDVEMANLTVSIILGFCHHQNDIC*F*NT*WQP*PLSKLHIKSLLELY 222
Query: 381 TAPEFFSPVGV---------------------PFDHSGDEQKSNVGLFQNQNAAQMTFSS 419
+ FS +G+ + EQ+SN GLFQN QM FS+
Sbjct: 223 LCIDEFS*LGLLVPLI*CYSFCML*LLQAIDAAYPMEAIEQRSNAGLFQNN--PQMAFST 396
Query: 420 DVSMGQVYNVVNDYEQPRNWNVATGGDTGIIIRARQAQNELVNTNI---NQGSANRRIRL 476
+V + QV VN EQ R+++ GDTGI IR RQ +N+ + N +QG A RRIRL
Sbjct: 397 NVGLDQVTCAVNYNEQSRSFDTVVHGDTGIRIRTRQGRNDQPDMNPTLPSQGIAPRRIRL 576
Query: 477 ---LKSAHGSSRVVKGGRRAQ-EDRNSKPITAVEKKASESLAADEDDTLTNHVKAMPKTP 532
+ +HGS K G A +D NSK I A EK+ASE+ AADE + K +
Sbjct: 577 GRLVMHSHGSDETAKDGSGAPPQDHNSKTIIAGEKEASENHAADESAAVDEPEKT---SS 747
Query: 533 KSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPCISKASSNRAM 575
+S ++RKI Q+ KDLLL RRV SK SS+RAM
Sbjct: 748 ESADSRKICQQ--PIAKPILGWKDLLLGRRVRYTSKVSSHRAM 870
>AV428075
Length = 310
Score = 105 bits (261), Expect = 3e-23
Identities = 49/97 (50%), Positives = 68/97 (69%), Gaps = 1/97 (1%)
Frame = +2
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGS-SLIGMKKTLVFYHGRAP 129
+ EW+F+ P+DRKY NG+R NR T GYWKATG DR I++ + IG+KKTLVFY G+AP
Sbjct: 5 EKEWYFYVPRDRKYRNGDRPNRVTTSGYWKATGADRMIRTENFRSIGLKKTLVFYSGKAP 184
Query: 130 KGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKK 166
KG RT+W+M+EYR E + + +CR++K+
Sbjct: 185 KGMRTSWIMNEYRLPQHETERYQKAE--ISLCRVYKR 289
>TC18524
Length = 452
Score = 98.6 bits (244), Expect = 3e-21
Identities = 53/72 (73%), Positives = 59/72 (81%)
Frame = +1
Query: 516 DEDDTLTNHVKAMPKTPKSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPCISKASSNRAM 575
DEDDTL+NHVKAM KTPKS +RKIY RGSNAGSS +VL DLLLLRRVPCISKASSNRA+
Sbjct: 1 DEDDTLSNHVKAMLKTPKSPAHRKIYPRGSNAGSSPQVLIDLLLLRRVPCISKASSNRAV 180
Query: 576 WSSVFVVSAFVM 587
VF +F +
Sbjct: 181 ---VFCFCSFCL 207
Score = 31.2 bits (69), Expect = 0.50
Identities = 14/17 (82%), Positives = 16/17 (93%)
Frame = +2
Query: 575 MWSSVFVVSAFVMVSVM 591
+WSSVFVVSAFVMV V+
Sbjct: 176 LWSSVFVVSAFVMVLVI 226
>TC12159 similar to UP|Q9M4U3 (Q9M4U3) NAC1, partial (21%)
Length = 630
Score = 95.9 bits (237), Expect = 2e-20
Identities = 43/85 (50%), Positives = 58/85 (67%)
Frame = +1
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
L GFRF P D E++ FYL++K+ G VI E++V K PWDLPD S +R D EW+
Sbjct: 376 LVPGFRFHPMDVELVKFYLKRKVMGRKLPHDVIAEVNVYKHAPWDLPDKSCLRTGDLEWY 555
Query: 76 FFCPQDRKYPNGNRLNRATILGYWK 100
FF P ++KY +G R+NRAT +G+WK
Sbjct: 556 FFSPTEKKYGSGARMNRATEIGFWK 630
>TC9181 similar to UP|Q9FY93 (Q9FY93) NAM-like protein
(AT5g13180/T19L5_140), partial (30%)
Length = 519
Score = 68.9 bits (167), Expect = 2e-12
Identities = 33/73 (45%), Positives = 47/73 (64%)
Frame = +1
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE++ YL++KI +I E+D+CK +PWDLP + + E +
Sbjct: 316 LPPGFRFHPTDEELVVQYLKRKIFSCPLPASIIPEVDLCKSDPWDLPG-----DSERERY 480
Query: 76 FFCPQDRKYPNGN 88
FF ++ KYPNGN
Sbjct: 481 FFSMKEAKYPNGN 519
>TC20162 similar to UP|Q8LKN6 (Q8LKN6) Nam-like protein 18, partial (17%)
Length = 623
Score = 60.8 bits (146), Expect = 6e-10
Identities = 25/34 (73%), Positives = 30/34 (87%)
Frame = +2
Query: 109 KSGSSLIGMKKTLVFYHGRAPKGKRTNWVMHEYR 142
K +L+GMKKTLVFY GRAPKG++TNWVMHE+R
Sbjct: 35 KGKGNLVGMKKTLVFYRGRAPKGEKTNWVMHEFR 136
>TC14586 similar to UP|Q9FY93 (Q9FY93) NAM-like protein
(AT5g13180/T19L5_140), partial (18%)
Length = 589
Score = 58.9 bits (141), Expect = 2e-09
Identities = 42/131 (32%), Positives = 62/131 (47%), Gaps = 10/131 (7%)
Frame = +3
Query: 115 IGMKKTLVFYHGRAPKGKRTNWVMHEYRPTLQELDGTNPG-----QNPYVICRLFKKQDE 169
+GMKKTLVFY G+ P G RT+W+MHEYR T G +V+CR+F K+
Sbjct: 3 VGMKKTLVFYTGKPPHGSRTDWIMHEYRLLTNTNTTTTQGHVQVPMENWVLCRIFLKRRG 182
Query: 170 SIEVSISGEAEQ-----TASTPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPE 224
++ +GE E + A + + + + + SSS T E
Sbjct: 183 GVK---NGEEEHSFNNISLKAGKAGRVVFYDFLAQNKINPASSSSSSATSGITDN---NE 344
Query: 225 TSEETTSNIIT 235
+ E +TSNI+T
Sbjct: 345 SDESSTSNILT 377
>AU252221
Length = 424
Score = 55.1 bits (131), Expect = 3e-08
Identities = 24/49 (48%), Positives = 35/49 (70%)
Frame = +1
Query: 14 NSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLP 62
++LP GFRF P+DEE+I YLR+K+ V +I E+D+ K +PW+LP
Sbjct: 100 SNLPPGFRFHPTDEELILHYLRKKVASIPLPVSIIAEVDIYKLDPWELP 246
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.131 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,212,863
Number of Sequences: 28460
Number of extensions: 121115
Number of successful extensions: 471
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 452
length of query: 596
length of database: 4,897,600
effective HSP length: 95
effective length of query: 501
effective length of database: 2,193,900
effective search space: 1099143900
effective search space used: 1099143900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0207.16


