

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.4
(368 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
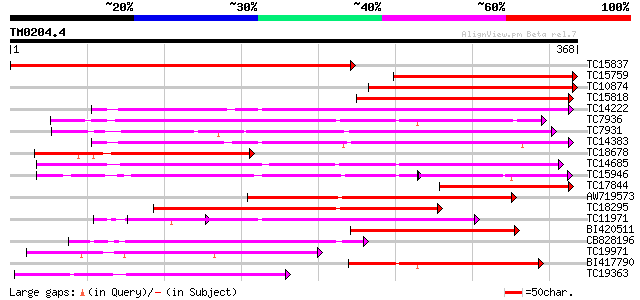
Score E
Sequences producing significant alignments: (bits) Value
TC15837 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyge... 469 e-133
TC15759 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxyge... 244 2e-65
TC10874 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxyge... 235 1e-62
TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyge... 233 3e-62
TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carb... 205 1e-53
TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, pa... 201 1e-52
TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial... 197 2e-51
TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-ca... 195 9e-51
TC18678 weakly similar to UP|Q9LY48 (Q9LY48) Leucoanthocyanidin ... 180 3e-46
TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 177 3e-45
TC15946 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 106 1e-35
TC17844 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxyge... 142 1e-34
AW719573 138 1e-33
TC18295 similar to UP|Q39224 (Q39224) SRG1 mRNA (F6I1.30/F6I1.30... 137 4e-33
TC11971 weakly similar to UP|Q43419 (Q43419) ACC oxidase, partia... 128 2e-30
BI420511 124 2e-29
CB828196 118 1e-27
TC19971 similar to UP|Q84V01 (Q84V01) Anthocyanidin synthase, pa... 118 2e-27
BI417790 116 5e-27
TC19363 similar to UP|Q39224 (Q39224) SRG1 mRNA (F6I1.30/F6I1.30... 114 3e-26
>TC15837 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (50%)
Length = 679
Score = 469 bits (1207), Expect = e-133
Identities = 224/224 (100%), Positives = 224/224 (100%)
Frame = +2
Query: 1 MMKKNISPLNWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDL 60
MMKKNISPLNWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDL
Sbjct: 8 MMKKNISPLNWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDL 187
Query: 61 RGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPME 120
RGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPME
Sbjct: 188 RGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPME 367
Query: 121 LKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFD 180
LKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFD
Sbjct: 368 LKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFD 547
Query: 181 EYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVN 224
EYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVN
Sbjct: 548 EYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVN 679
>TC15759 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxygenase-like
protein, partial (34%)
Length = 929
Score = 244 bits (622), Expect = 2e-65
Identities = 119/119 (100%), Positives = 119/119 (100%)
Frame = +2
Query: 250 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 309
LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV
Sbjct: 2 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 181
Query: 310 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 368
SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR
Sbjct: 182 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 358
>TC10874 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxygenase-like
protein, partial (38%)
Length = 723
Score = 235 bits (599), Expect = 1e-62
Identities = 116/135 (85%), Positives = 122/135 (89%)
Frame = +2
Query: 234 LTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIY 293
LTLGLSSHSDPGGMT+LLPDDQV GLQVRKGD+WITV P HAFIVNIGDQIQVLSNAIY
Sbjct: 2 LTLGLSSHSDPGGMTMLLPDDQVRGLQVRKGDDWITVNPARHAFIVNIGDQIQVLSNAIY 181
Query: 294 KSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMK 353
SVEHRVIVNS+KERVSLAFFYNP+ DIPIEP KELV DKPALY AMTFDEYR FIRM+
Sbjct: 182 TSVEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYRLFIRMR 361
Query: 354 GPCGKSHVESLKSPR 368
GP GKS VES+KSPR
Sbjct: 362 GPRGKSQVESMKSPR 406
>TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (37%)
Length = 618
Score = 233 bits (595), Expect = 3e-62
Identities = 110/141 (78%), Positives = 127/141 (90%)
Frame = +2
Query: 226 YPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQI 285
YPKCP+P+LTLGLS HSDPGG+T+LLPDD V+GLQVRKG+ WITVKPVP+AFI+NIGDQI
Sbjct: 2 YPKCPQPDLTLGLSPHSDPGGLTILLPDDFVSGLQVRKGNYWITVKPVPNAFIINIGDQI 181
Query: 286 QVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDE 345
QVLSN IYKSVEHRVIVNSN++RVSLA FYNP+ D+ IEPAKELV E+KPALY MT+DE
Sbjct: 182 QVLSNTIYKSVEHRVIVNSNQDRVSLAMFYNPKSDLLIEPAKELVTEEKPALYPPMTYDE 361
Query: 346 YRRFIRMKGPCGKSHVESLKS 366
YR +IR +GPCGK+ VESL S
Sbjct: 362 YRLYIRRQGPCGKAQVESLAS 424
>TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carboxylate
oxidase homolog (ACC oxidase) , partial (97%)
Length = 1597
Score = 205 bits (521), Expect = 1e-53
Identities = 117/316 (37%), Positives = 182/316 (57%), Gaps = 3/316 (0%)
Frame = +1
Query: 54 NIPIIDLRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRK 113
N P++D+ L + E R +T+ I +AC+ WGFF++VNH +S + MD ++
Sbjct: 52 NFPVVDMSKL-------NTEERVATMELIKDACENWGFFELVNHEISTEFMDTVERLTKE 210
Query: 114 FFHLPMELKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPP 173
+ ME + + + K E S + LDW +FL +LP S + ++ P L
Sbjct: 211 HYKRFMEQRFKEMVATKGLETVQSEIDD-----LDWESTFFLRHLPSS--NISEVPDLDE 369
Query: 174 SCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGF-GGEDIGACLRVNFYPKCPKP 232
R+V E+ +L KL L+ +L NL LE+ +L+ F G + +V+ YP CPKP
Sbjct: 370 DYRKVMKEFAVKLEKLAENLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKP 549
Query: 233 ELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAI 292
+L GL +H+D GG+ LL DD+V+GLQ+ K D WI V P+ H+ +VN+GDQ++V++N
Sbjct: 550 DLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMRHSIVVNLGDQLEVITNGK 729
Query: 293 YKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPA-LYTAMTFDEYRR-FI 350
YKSV HRVI ++ R+SLA FYNP D I PA LVKED+ + +Y F++Y + ++
Sbjct: 730 YKSVMHRVIAQTDGARMSLASFYNPGDDAVISPAPSLVKEDETSQVYPKFVFEDYMKLYL 909
Query: 351 RMKGPCGKSHVESLKS 366
+K + E +K+
Sbjct: 910 GVKFQAKEPRFEVMKA 957
>TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(94%)
Length = 1492
Score = 201 bits (512), Expect = 1e-52
Identities = 118/326 (36%), Positives = 187/326 (57%), Gaps = 4/326 (1%)
Frame = +3
Query: 27 DSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEAC 86
+++ +++ +RP + ++ S N IP+I L G+ + + R+ +I EAC
Sbjct: 186 NTLESSFVRDEDERPKVAYNNFS---NEIPVISLAGI-----DEVDDRRSEICNKIVEAC 341
Query: 87 KEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGSRLGVEKGAI 146
+ WG FQ+V+HGV +L+ ++FF LP E K ++ + G+ ++ ++
Sbjct: 342 ENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESV 521
Query: 147 LDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEED 206
DW + P+ +DY++WP P + V +EY +L+ L +L++VLS + LE++
Sbjct: 522 QDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKE 701
Query: 207 FLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRK--G 264
L D+ + VN+YPKCP+P+LTLGL H+DPG +TLLL DQV GLQ + G
Sbjct: 702 ALTKAC--VDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLL-QDQVGGLQATRDNG 872
Query: 265 DNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIE 324
WITV+PV AF+VN+GD LSN +K+ +H+ +VNSN R+S+A F NP D +
Sbjct: 873 KTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVY 1052
Query: 325 PAKELVKE-DKPALYTAMTFDE-YRR 348
P K V+E +K + +TF E YRR
Sbjct: 1053PLK--VREGEKSVMEEPITFAEMYRR 1124
>TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial (29%)
Length = 1477
Score = 197 bits (501), Expect = 2e-51
Identities = 116/332 (34%), Positives = 183/332 (54%), Gaps = 4/332 (1%)
Frame = +1
Query: 28 SIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEACK 87
++P Y++P RP + +S +P++DL G RA TL+QI +A +
Sbjct: 61 TVPLSYVQPPESRPGTVTVASGKA---VPVVDLGG----------HDRAETLKQIFKASE 201
Query: 88 EWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEG----YGSRLGVEK 143
E+GFFQ++NHGVS +L++ +++F +P E ++ + S + G Y SR K
Sbjct: 202 EYGFFQVINHGVSNELIEDTLNIFKEFHAMPAE--EKLSESSRDPNGSCMLYTSREINNK 375
Query: 144 GAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLEL 203
I W D H S + WP P REV ++Y +EL +L +++++L L L
Sbjct: 376 DCIQFWKDT-LRHPCTPSEEFMEFWPLKPARYREVVEKYTKELRELGSQILEMLCEGLGL 552
Query: 204 EEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRK 263
+ + G + L N YP CP+P LTLGLS H DP +T+LL D V LQV K
Sbjct: 553 DPRYCCAGLSENPL---LLSNHYPPCPEPSLTLGLSKHRDPTLVTILLQDTDVNALQVFK 723
Query: 264 GDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPI 323
WI+++P+P+AF+VNIG +Q++SN VEHRV+ NS+ R ++A+F P+ + +
Sbjct: 724 DGEWISIEPIPYAFVVNIGLLLQIISNGRLVGVEHRVVTNSSISRTTVAYFIRPKKEEIV 903
Query: 324 EPAKELVKEDKPALYTAMTFDEYRRFIRMKGP 355
EPAK LV+ +Y ++ F+E+ R + + P
Sbjct: 904 EPAKALVRTGAHPIYRSIAFEEFLRIFKTRDP 999
>TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1249
Score = 195 bits (496), Expect = 9e-51
Identities = 114/320 (35%), Positives = 177/320 (54%), Gaps = 7/320 (2%)
Frame = +3
Query: 54 NIPIIDLRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRK 113
N P+I L L + E R QI +AC+ WGFF++VNHG+ D+MD ++
Sbjct: 45 NFPVISLEKL-------NGEERKDIKLQIKDACENWGFFELVNHGIPHDVMDTVERLTKE 203
Query: 114 FFHLPMELKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPP 173
+ + ME + + + K E + + +DW + L +LP S + ++ P L
Sbjct: 204 HYRICMEQRFKELMASKGLEAVQTEV-----KDMDWESTFHLRHLPES--NISEIPDLTD 362
Query: 174 SCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGE---DIGACLRVNFYPKCP 230
R+V ++ + KL L+ +L NL LE+ +L+ F G G +V YP CP
Sbjct: 363 EYRKVMKDFALRIEKLSEDLLDLLCENLGLEKGYLKKAFHGSRGPTFGT--KVANYPPCP 536
Query: 231 KPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSN 290
KP L GL +H+D GG+ LL DD+V+GLQ+ K W+ V P+ H+ +VN+GDQ++V++N
Sbjct: 537 KPNLVKGLRAHTDAGGIILLFQDDKVSGLQLLKDGEWVDVPPMRHSIVVNLGDQLEVITN 716
Query: 291 AIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVK---EDKPALYTAMTFDEYR 347
YKSVEHRVI ++ R+S+A FYNP D I PA EL++ E+K LY F++Y
Sbjct: 717 GKYKSVEHRVIAQTDGTRMSIASFYNPGSDAVIYPAPELLEKEAEEKNQLYPKFVFEDYM 896
Query: 348 R-FIRMKGPCGKSHVESLKS 366
+ + +K + E+LK+
Sbjct: 897 KLYAGLKFDAKEPRFEALKA 956
>TC18678 weakly similar to UP|Q9LY48 (Q9LY48) Leucoanthocyanidin
dioxygenase-like protein, partial (28%)
Length = 451
Score = 180 bits (457), Expect = 3e-46
Identities = 90/151 (59%), Positives = 112/151 (73%), Gaps = 8/151 (5%)
Frame = +3
Query: 17 RVQSLSETCIDSIPERYIKPLTDRPSI------NSSSSSSDAN--NIPIIDLRGLLYHGD 68
RVQ+L+E+ I SIPER+IKP + RP+ ++S D N NIP+IDL+ H
Sbjct: 6 RVQALAESGISSIPERFIKPTSQRPTKTTFTNNHASEVHDDENTINIPVIDLQ----HVY 173
Query: 69 PDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANS 128
D TL+++SEAC+EWGFFQ+VNHGVS +LM ARE WR+FFHLP E+K+ YANS
Sbjct: 174 GDDSRLCEETLKRVSEACREWGFFQVVNHGVSHELMKHAREVWREFFHLPHEVKEDYANS 353
Query: 129 PKTYEGYGSRLGVEKGAILDWSDYYFLHYLP 159
P TYEGYGSRLGV+KGAILDWSDY+FLHY+P
Sbjct: 354 PTTYEGYGSRLGVKKGAILDWSDYFFLHYMP 446
>TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (38%)
Length = 1323
Score = 177 bits (448), Expect = 3e-45
Identities = 119/345 (34%), Positives = 176/345 (50%), Gaps = 3/345 (0%)
Frame = +1
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
V+ L ++ I +IP ++ P + S IP IDL + E+RA+
Sbjct: 187 VKGLIDSGIKTIPSFFVHPPETLSDLKPRPESDSKPEIPTIDLAAV--------NESRAA 342
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQ-YANSPKTYEGYG 136
+ QI A GFFQ+VNHGV DL+ + F P E + + Y T Y
Sbjct: 343 VVDQIRRAASSVGFFQVVNHGVPLDLLRRTVAAIKAFHEQPEEERARVYTREMGTGVSYI 522
Query: 137 SRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKV 196
S + + + W D L P ++ +P CR+ E+ +E+V++ L +
Sbjct: 523 SNVDLFQSKAASWRDTLQLRMGPTAVDQ----SMIPEVCRQEVMEWDKEVVRVGEVLYGL 690
Query: 197 LSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQV 256
LS L L+ L G G + ++YP CP+P+LT+GL+SH+DPG +T++L D +
Sbjct: 691 LSEGLGLDAGRLSEM--GLVQGRVMVGHYYPFCPQPDLTVGLNSHADPGALTVVL-QDHI 861
Query: 257 TGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNS-NKERVSLAFFY 315
GLQVR + W+ VKP+ A ++NIGD +Q++SN YKS +HRV+ NS N+ RVS+A F
Sbjct: 862 GGLQVRTTEGWVDVKPLDGALVINIGDLLQIISNEEYKSADHRVLANSANEPRVSVAVFL 1041
Query: 316 NP-RGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKS 359
NP + P EL +KPALY T DE+ K GKS
Sbjct: 1042NPGNREKLFGPLPELTSAEKPALYRNFTLDEFLTRFFKKELDGKS 1176
>TC15946 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (56%)
Length = 1252
Score = 106 bits (264), Expect(2) = 1e-35
Identities = 70/252 (27%), Positives = 127/252 (49%), Gaps = 1/252 (0%)
Frame = +1
Query: 18 VQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAS 77
V+ L E + IP + ++ + +S +++P+IDL +H E
Sbjct: 130 VKGLVECGVTKIPRMFHA----NKHLDITPENSKLSSVPLIDLTD-------EHSEV--- 267
Query: 78 TLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQY-ANSPKTYEGYG 136
+ +I AC EWGFFQ++NHG+ ++D + R+F E+++Q+ + K Y
Sbjct: 268 -IGKIRSACHEWGFFQVINHGIPTSVLDEMIDGIRRFHEQDSEVRKQFHSRDLKKKVMYY 444
Query: 137 SRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKV 196
+ + + G +W D + P D K +P CR++ EY +++ +L ++++
Sbjct: 445 TNISLFSGQAANWRDTFGFAVAP----DPPKPDEIPLVCRDIVMEYSKKIKELGFTILEL 612
Query: 197 LSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQV 256
LS L L +L+ E G + ++YP CP+P+LT+G + H+D +TLLL DQ+
Sbjct: 613 LSEGLGLNPSYLKELNCAE--GLFVLGHYYPPCPEPKLTMGTTKHTDGNFITLLL-QDQL 783
Query: 257 TGLQVRKGDNWI 268
GLQ+ + W+
Sbjct: 784 GGLQILHENQWV 819
Score = 60.1 bits (144), Expect(2) = 1e-35
Identities = 38/107 (35%), Positives = 57/107 (52%), Gaps = 7/107 (6%)
Frame = +2
Query: 266 NWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIE- 324
N V PV A +VNIGD +Q+++N + SV HRV+ + R+S+A F+ D PIE
Sbjct: 812 NGXDVHPVHGALVVNIGDLLQLITNDRFVSVYHRVLSQNIGPRISVASFFVNSPD-PIEG 988
Query: 325 ------PAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
P KEL+ E+ P +Y +T ++ KG G S +E +
Sbjct: 989 TLKVYGPIKELLTEENPPIYRDVTIKDFLAHYYAKGLDGNSSLEPFR 1129
>TC17844 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxygenase-like
protein, partial (25%)
Length = 597
Score = 142 bits (357), Expect = 1e-34
Identities = 68/87 (78%), Positives = 78/87 (89%)
Frame = +1
Query: 280 NIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALYT 339
NIGDQIQVLSNAIYKS+EHRV+VNSNK+RVSLAFFYNPR D+ I+PA+ELV +D+PALY
Sbjct: 1 NIGDQIQVLSNAIYKSIEHRVMVNSNKDRVSLAFFYNPRSDLLIKPAEELVTKDRPALYP 180
Query: 340 AMTFDEYRRFIRMKGPCGKSHVESLKS 366
MTFDEYR +IR KGPCGK+ VESL S
Sbjct: 181 PMTFDEYRLYIRTKGPCGKAQVESLIS 261
>AW719573
Length = 521
Score = 138 bits (348), Expect = 1e-33
Identities = 67/175 (38%), Positives = 109/175 (62%)
Frame = +2
Query: 155 LHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGG 214
L LP ++ + + +P RE + Y EL L ++M++++ L+++ + +
Sbjct: 2 LRTLPPHIRKPHLFNHIPQPFRENLEIYCAELNNLSIKIMELMANALKIDPKEITEIYN- 178
Query: 215 EDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVP 274
+ +R+N+YP CP+PE +GL SH+D G+T+LL + + GLQ+RK +W+ V+P+P
Sbjct: 179 -EGSQTIRMNYYPPCPQPERVIGLKSHTDGAGLTILLQANDIEGLQIRKDGHWVPVQPLP 355
Query: 275 HAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKEL 329
+AFI+NIGD +++ +N IY S+EHR VNS KER+SLA FY P + PA L
Sbjct: 356 NAFIINIGDMLEITTNGIYASIEHRATVNSEKERISLATFYGPNMQATLAPAPSL 520
>TC18295 similar to UP|Q39224 (Q39224) SRG1 mRNA (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (29%)
Length = 561
Score = 137 bits (344), Expect = 4e-33
Identities = 63/188 (33%), Positives = 117/188 (61%)
Frame = +3
Query: 94 IVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGSRLGVEKGAILDWSDYY 153
++NHGV+P L++ ++ + F+LP+E K++ +P+ +G+G V + L+W+D +
Sbjct: 3 LINHGVNPSLVENMKKGVQDLFNLPIEEKKKLWQTPEVGQGFGQAFVVSEEQKLEWADLF 182
Query: 154 FLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFG 213
+++ PL+ ++ + P++P R+ D Y EL +C ++ ++ L+ E + L F
Sbjct: 183 YINTFPLNKRNTSLIPSIPQPFRDNLDAYCSELKNICVTIIGLMEKALKTETNELVELF- 359
Query: 214 GEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPV 273
ED +R+N+YP CP+PE +GL+ HSD G T+LL +++ GLQ+RK W+ +KP+
Sbjct: 360 -EDTSQGMRMNYYPPCPQPEKVIGLNPHSDSGAFTILLQVNEMEGLQIRKDGIWVPIKPL 536
Query: 274 PHAFIVNI 281
+AF++NI
Sbjct: 537 SNAFVINI 560
>TC11971 weakly similar to UP|Q43419 (Q43419) ACC oxidase, partial (69%)
Length = 730
Score = 128 bits (321), Expect = 2e-30
Identities = 80/235 (34%), Positives = 128/235 (54%), Gaps = 6/235 (2%)
Frame = +3
Query: 77 STLRQISEACKEWGF-FQIVNHGVSPDL--MDMARETWRKFFH-LPMELKQQYANSPKTY 132
S L ++ K W F ++V G + L M++ R WR+ L + +++ + + +
Sbjct: 24 SALSMETKGVKPWHFCMRLVRTGGAF*LRTMELTRALWRR*SS*LTLTMRKT*SKAFTSL 203
Query: 133 EGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGR 192
+ +K + +DW +F+ + P S + + L R+ D+Y +L++L
Sbjct: 204 RKQRAWRKKQKASDIDWESAFFIWHRPKS--NIKQISNLSEELRQTMDQYIEKLIQLAED 377
Query: 193 LMKVLSLNLELEEDFLQNGFGGEDIGAC-LRVNFYPKCPKPELTLGLSSHSDPGGMTLLL 251
L ++S NL LE+D+++ F G D A +V YP+CP PEL GL H+D GG+ LLL
Sbjct: 378 LSGLMSENLGLEKDYIKKAFSGSDGPAMGTKVAKYPQCPNPELVRGLREHTDAGGIILLL 557
Query: 252 PDDQVTGLQVRKGDNWITVKPVP-HAFIVNIGDQIQVLSNAIYKSVEHRVIVNSN 305
DDQV GL+ K W+ + P +A +N GDQI+VLSN +YKSV HRV+ + N
Sbjct: 558 QDDQVPGLEFSKDGKWVEIPPSKNNAIFINTGDQIEVLSNGLYKSVVHRVMADKN 722
Score = 46.2 bits (108), Expect = 9e-06
Identities = 29/76 (38%), Positives = 37/76 (48%)
Frame = +2
Query: 55 IPIIDLRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKF 114
IPIID L +GD R T+ + EACK WG F I NHG+ LM+ ++
Sbjct: 8 IPIIDFSTL--NGDK-----RGETMALLHEACKNWGCFLIENHGIDKSLMEKVKQLINT- 163
Query: 115 FHLPMELKQQYANSPK 130
H LKQ + S K
Sbjct: 164 -HYEENLKQSFYESEK 208
>BI420511
Length = 384
Score = 124 bits (311), Expect = 2e-29
Identities = 56/110 (50%), Positives = 79/110 (70%)
Frame = +3
Query: 222 RVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNI 281
+V YP CPKPEL GL +H+D GG+ LLL DD+V+GLQ+ K W+ V P+ H+ ++N+
Sbjct: 24 KVANYPACPKPELVKGLRAHTDAGGIILLLQDDKVSGLQLLKDGQWVDVPPMRHSIVINL 203
Query: 282 GDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVK 331
GDQ++V++N YKSVEHRVI ++ R+S+A FYNP D I PA L++
Sbjct: 204 GDQLEVITNGKYKSVEHRVIAKTDGTRMSIASFYNPANDAVIYPAPALLE 353
>CB828196
Length = 555
Score = 118 bits (296), Expect = 1e-27
Identities = 66/195 (33%), Positives = 109/195 (55%)
Frame = +3
Query: 39 DRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHG 98
+RP++ +SS++ +PIIDL LL DH + L ++ +ACKEWGFFQ++NHG
Sbjct: 3 ERPALYNSSTTPLP--LPIIDLSKLL---SKDH---KVPELERLHQACKEWGFFQLINHG 158
Query: 99 VSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYL 158
V+ L++ + ++FFHLPME K+++ EGYG V + L+W D +F+
Sbjct: 159 VNTSLLEDVKRGVQEFFHLPMEEKKKFEQRQGDAEGYGQVFVVSEDQKLEWGDVFFMFLF 338
Query: 159 PLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGEDIG 218
PL + +P LP R+ D Y E+ KL ++++++ L+++ + D
Sbjct: 339 PLEKRKPYLFPKLPSKFRDDLDTYYTEVKKLGIHILELMANALKIDTKEITELL---DTT 509
Query: 219 ACLRVNFYPKCPKPE 233
R+N+YP CP+PE
Sbjct: 510 QATRLNYYPPCPQPE 554
>TC19971 similar to UP|Q84V01 (Q84V01) Anthocyanidin synthase, partial (53%)
Length = 595
Score = 118 bits (295), Expect = 2e-27
Identities = 67/198 (33%), Positives = 103/198 (51%), Gaps = 6/198 (3%)
Frame = +1
Query: 12 PEPIVRVQSLSETCIDSIPERYIKPLTDRPSINS--SSSSSDANNIPIIDLRGLLYHGDP 69
P + RV+SLS + I SIP+ Y++P + +I +P IDL+ D
Sbjct: 19 PTVVERVESLSGSGIQSIPKEYVRPKEELANIGDVFEEEKKVGPQVPTIDLK------DI 180
Query: 70 DHPE--ARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYAN 127
D P+ RA ++ +A +EWG +VNHG+ +L++ + +FF LP+E K++YAN
Sbjct: 181 DSPDEFVRAKCREKLRKAAEEWGVMHLVNHGIPDELLNQLKSAGAEFFSLPVEEKEKYAN 360
Query: 128 SPKT--YEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRE 185
+GYGS+L L+W DY+F P +D + WP P EV +Y R
Sbjct: 361 DQAAGNVQGYGSKLANNASGQLEWEDYFFHLIFPEDKRDLSIWPKTPSYYTEVTSDYARR 540
Query: 186 LVKLCGRLMKVLSLNLEL 203
L L ++++VLSL L L
Sbjct: 541 LRVLASKILEVLSLELGL 594
>BI417790
Length = 610
Score = 116 bits (291), Expect = 5e-27
Identities = 55/128 (42%), Positives = 80/128 (61%), Gaps = 2/128 (1%)
Frame = +3
Query: 221 LRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRK--GDNWITVKPVPHAFI 278
+R+N+YP CP P++ LG H D G +T+L D+V+GLQVR+ W+ + P+P A+I
Sbjct: 6 IRLNYYPPCPNPDIVLGCGRHKDSGALTILA-QDEVSGLQVRRKCDGQWVRLNPLPDAYI 182
Query: 279 VNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALY 338
+N+GD IQV N Y+SVEHRV +N K R+S FF P +EP +EL E+ P Y
Sbjct: 183 INVGDVIQVWCNDAYESVEHRVKLNPEKARLSYPFFLYPSHYTNVEPLEELTNEENPPKY 362
Query: 339 TAMTFDEY 346
T + ++
Sbjct: 363 TPYNWGKF 386
>TC19363 similar to UP|Q39224 (Q39224) SRG1 mRNA (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (23%)
Length = 552
Score = 114 bits (284), Expect = 3e-26
Identities = 59/179 (32%), Positives = 102/179 (56%)
Frame = +2
Query: 4 KNISPLNWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGL 63
+ +S L + VQ L++ + +PE+Y++P D IN + +S+ + +P+IDL L
Sbjct: 41 RKMSKLGTSLLVPSVQELAKQHMIELPEQYLRPNQD--PINVAPNSTSSPQVPVIDLNKL 214
Query: 64 LYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQ 123
L A+ L+++ ACKEWGFFQ++NHGV P L++ + + FF LPME K+
Sbjct: 215 LSED--------ATELQKLHSACKEWGFFQLINHGVKPSLVENVKIGVQDFFSLPMEEKK 370
Query: 124 QYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEY 182
++ +P+ EG+G V + LDW+D +F++ LP ++ +P +P R+ + Y
Sbjct: 371 KFWQTPEDIEGFGQAFVVSEDQKLDWADLFFINTLPSDARNPRLFPNIPQPLRDNVESY 547
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,260,523
Number of Sequences: 28460
Number of extensions: 112553
Number of successful extensions: 692
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 646
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 651
length of query: 368
length of database: 4,897,600
effective HSP length: 92
effective length of query: 276
effective length of database: 2,279,280
effective search space: 629081280
effective search space used: 629081280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0204.4


