

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.2
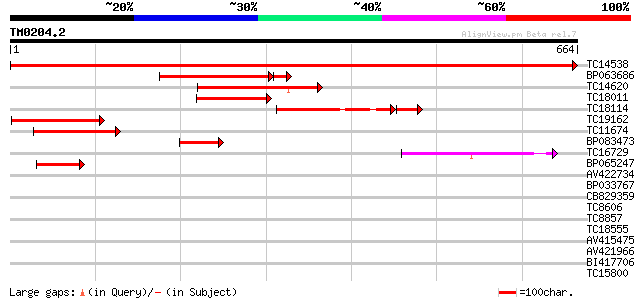
(664 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14538 similar to GB|AAH54382.1|32450414|BC054382 feminization ... 1336 0.0
BP063686 221 4e-63
TC14620 166 1e-41
TC18011 152 2e-37
TC18114 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D... 123 4e-34
TC19162 105 2e-23
TC11674 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D... 100 1e-21
BP083473 100 1e-21
TC16729 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D... 88 4e-18
BP065247 66 2e-11
AV422734 37 0.008
BP033767 34 0.086
CB829359 33 0.15
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 33 0.19
TC8857 similar to UP|Q8LKG0 (Q8LKG0) Drm4, partial (84%) 32 0.25
TC18555 similar to GB|AAP68307.1|31711902|BT008868 At2g33570 {Ar... 32 0.33
AV415475 31 0.56
AV421966 31 0.56
BI417706 31 0.73
TC15800 homologue to GB|AAM19932.1|20453387|AY097416 AT4g17800/d... 31 0.73
>TC14538 similar to GB|AAH54382.1|32450414|BC054382 feminization 1 homolog a
{Mus musculus;}, partial (3%)
Length = 2631
Score = 1336 bits (3458), Expect = 0.0
Identities = 664/665 (99%), Positives = 664/665 (99%), Gaps = 1/665 (0%)
Frame = +2
Query: 1 MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF 60
MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF
Sbjct: 101 MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF 280
Query: 61 WYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAG 120
WYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAG
Sbjct: 281 WYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAG 460
Query: 121 GSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQL 180
GSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQL
Sbjct: 461 GSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQL 640
Query: 181 IHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSH 240
IHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSH
Sbjct: 641 IHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSH 820
Query: 241 DWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYR 300
DWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYR
Sbjct: 821 DWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYR 1000
Query: 301 TRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSS 360
TRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSS
Sbjct: 1001TRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSS 1180
Query: 361 LPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEME 420
LPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEME
Sbjct: 1181LPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEME 1360
Query: 421 MLGLGSPTR-QQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQL 479
MLGLGSPTR QQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQL
Sbjct: 1361MLGLGSPTRQQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQL 1540
Query: 480 HGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVM 539
HGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVM
Sbjct: 1541HGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVM 1720
Query: 540 NSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDEL 599
NSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDEL
Sbjct: 1721NSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDEL 1900
Query: 600 SKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDLSKEMLPPWVEQLYIEQ 659
SKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDLSKEMLPPWVEQLYIEQ
Sbjct: 1901SKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDLSKEMLPPWVEQLYIEQ 2080
Query: 660 EQMVA 664
EQMVA
Sbjct: 2081EQMVA 2095
>BP063686
Length = 514
Score = 221 bits (562), Expect(2) = 4e-63
Identities = 98/133 (73%), Positives = 111/133 (82%)
Frame = -2
Query: 176 ESGQLIHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCS 235
E+G ++ L+ S+ + S+KKEYPVD SLPDI N +Y +DEFRMYSFKV+PCS
Sbjct: 486 ENGSPSALDLQLKSKSSDVPAFSVSEKKEYPVDPSLPDIKNSIYSTDEFRMYSFKVRPCS 307
Query: 236 RAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLH 295
RAYSHDWTECPFVHPGENARRRDPRKY YSCVPCP+FRKGAC++GD CEYAHGVFE WLH
Sbjct: 306 RAYSHDWTECPFVHPGENARRRDPRKYHYSCVPCPDFRKGACRRGDMCEYAHGVFECWLH 127
Query: 296 PAQYRTRLCKDET 308
PAQYRTRLCKD T
Sbjct: 126 PAQYRTRLCKDGT 88
Score = 38.5 bits (88), Expect(2) = 4e-63
Identities = 14/21 (66%), Positives = 17/21 (80%)
Frame = -1
Query: 310 CGRKVCFFAHRPEELRPVYAS 330
C R++CFFAH EELRP+Y S
Sbjct: 85 CSRRICFFAHTAEELRPLYVS 23
>TC14620
Length = 1260
Score = 166 bits (420), Expect = 1e-41
Identities = 75/151 (49%), Positives = 97/151 (63%), Gaps = 5/151 (3%)
Frame = +3
Query: 221 SDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKG 280
+D FRM+ FKV C R HDWT+CP+ HPGE ARRRDPRK+ YS PCP+ RKG C +G
Sbjct: 165 TDHFRMFDFKVNLCQRGRPHDWTDCPYAHPGEKARRRDPRKHHYSATPCPDLRKGYCNRG 344
Query: 281 DSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELR-----PVYASTGSAM 335
DSC +AHGVFE WLHP++YRT+ CKD T C R+VCFFAH +LR V +S S +
Sbjct: 345 DSCHFAHGVFECWLHPSRYRTQPCKDGTFCRRRVCFFAHTANQLRVLNHEHVLSSPTSVL 524
Query: 336 PSPKSYSSNAVDMSVMSPLALSSSSLPMSTV 366
S + ++ + V+S ++ M V
Sbjct: 525 NSSSTLTTKSFRYGVVSAFSVDEIVRSMRNV 617
>TC18011
Length = 542
Score = 152 bits (383), Expect = 2e-37
Identities = 62/88 (70%), Positives = 72/88 (81%)
Frame = +3
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
Y D FRMY FKV+ C+R SHDWTECP+ HPGE ARRRDPRK+ YS CP+FRKG+C+
Sbjct: 276 YSCDHFRMYEFKVRRCARGRSHDWTECPYAHPGEKARRRDPRKFHYSGTACPDFRKGSCK 455
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKD 306
K D+CEYAHGVFE WLHPA+YRT+ CKD
Sbjct: 456 KADACEYAHGVFECWLHPARYRTQPCKD 539
>TC18114 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;}, partial (4%)
Length = 576
Score = 123 bits (309), Expect(2) = 4e-34
Identities = 68/141 (48%), Positives = 93/141 (65%), Gaps = 2/141 (1%)
Frame = +2
Query: 313 KVCFFAHRPEELRPVYASTGSAMPSPKSYSSN--AVDMSVMSPLALSSSSLPMSTVSTPP 370
+VCFFAH+ EELRP+Y STGSA+PSP+SYS+N A++M +SP+A S S M VSTPP
Sbjct: 2 RVCFFAHKLEELRPLYVSTGSAIPSPRSYSANNSALEMGSVSPIAFGSPSSMMPPVSTPP 181
Query: 371 MSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQ 430
+SP S G +WQ N++ P+LQLP SRL TA +ARD D E L R
Sbjct: 182 LSPSGASSPVAGGAMWQ-----NVSVPALQLPKSRLNTAFAARDIDFHNEFL------RL 328
Query: 431 QQQQQQQQQLIEEIARISSPS 451
+ +++QQ + +E++ +SSPS
Sbjct: 329 EINRRRQQLMKDEVSGLSSPS 391
Score = 38.9 bits (89), Expect(2) = 4e-34
Identities = 15/30 (50%), Positives = 23/30 (76%)
Frame = +3
Query: 454 NRIGDLTPTNLDDLLGSVDSNLLSQLHGLS 483
NR+ + P N +D+ GS+D ++L+QLHGLS
Sbjct: 462 NRLSGVNPANFEDMYGSLDRSVLAQLHGLS 551
>TC19162
Length = 438
Score = 105 bits (263), Expect = 2e-23
Identities = 57/110 (51%), Positives = 76/110 (68%), Gaps = 1/110 (0%)
Frame = +2
Query: 3 SGSKSKLSSSEQVMESQKENCDGLHN-FSILLELSASNDFEALKREVDEKGLDVNEAGFW 61
SGS K S S VME++ + LH+ S LLE SA++D K V+++ DV+EAG W
Sbjct: 107 SGSDKKASQSGFVMENEYGKQEELHHKISALLEFSAADDLIGFKDAVEKEDHDVDEAGLW 286
Query: 62 YGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKA 111
YGRR+GS K+G E+RTPLM+A++FGS V Y++ TGRVDVN+A SD A
Sbjct: 287 YGRRVGSNKLGYEERTPLMVAAMFGSLGVSIYILGTGRVDVNRASRSDGA 436
>TC11674 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;}, partial (14%)
Length = 570
Score = 100 bits (248), Expect = 1e-21
Identities = 48/102 (47%), Positives = 70/102 (68%)
Frame = +3
Query: 28 NFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGS 87
+FS LLEL+++NDFE K + + +NE G WY R+ GSK++ E RTPLM+A+ +GS
Sbjct: 264 SFSSLLELASNNDFEGFKELLYKDASSINEVGLWYVRQNGSKQIVLEHRTPLMVAATYGS 443
Query: 88 TRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVV 129
V+K ++ DVN ACG +K+TALHCA +GGS +++ V
Sbjct: 444 IDVLKVILSCPEADVNFACGVNKSTALHCAASGGSVKAVDAV 569
>BP083473
Length = 214
Score = 99.8 bits (247), Expect = 1e-21
Identities = 40/51 (78%), Positives = 47/51 (91%)
Frame = -3
Query: 200 SDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHP 250
++KKEYP+D SLPDI N +Y +DEFRM+SFKV+PCSRAYSHDWTECPFVHP
Sbjct: 155 AEKKEYPIDPSLPDIKNSIYATDEFRMFSFKVRPCSRAYSHDWTECPFVHP 3
>TC16729 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;}, partial (8%)
Length = 1180
Score = 88.2 bits (217), Expect = 4e-18
Identities = 59/191 (30%), Positives = 99/191 (50%), Gaps = 8/191 (4%)
Frame = +1
Query: 459 LTPTNLDDLLGS-VDSNLLSQLHGLSGPSTPTHLQSQ-SRHQMMQQNMNHLRSSYPSNIP 516
LTP+NLDDL + + S+ + +PTH + ++ Q +Q +++ + ++ S
Sbjct: 58 LTPSNLDDLFSAEISSSPRYSDPAAASVFSPTHRSAVLNQFQQLQSSLSPINTNVLSPNN 237
Query: 517 SSPVRKPSSLGFDSSSAVAAAVM------NSRSAAFAKRSQSFIDRGAAGTHHLGMSSPS 570
+ SS G S ++ + +SR +AFA+R + + + LG ++P+
Sbjct: 238 GEHPLRDSSFGVSSPGRMSPRSVEPLSPGSSRLSAFAQREKQQQQLRSLSSRDLGANNPA 417
Query: 571 NPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEP 630
+ + +S W SP+GK+DW VNG+EL +L++S+SF NNG EP
Sbjct: 418 SIVGSPINSWSKWGSPTGKVDWSVNGNELGRLQRSSSFELGNNG-------------EEP 558
Query: 631 DVSWVHSLVKD 641
D+SWV SLVKD
Sbjct: 559 DLSWVQSLVKD 591
>BP065247
Length = 528
Score = 66.2 bits (160), Expect = 2e-11
Identities = 31/56 (55%), Positives = 42/56 (74%)
Frame = +3
Query: 32 LLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGS 87
LLEL+A+ND E KR ++ V+E G WYGRR GSK+M +E+RTPLM+A+ +GS
Sbjct: 360 LLELAANNDVEGFKRLIECDPSSVDEVGLWYGRRKGSKQMVNEQRTPLMVAATYGS 527
>AV422734
Length = 448
Score = 37.4 bits (85), Expect = 0.008
Identities = 28/86 (32%), Positives = 39/86 (44%)
Frame = +1
Query: 429 RQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSGPSTP 488
+Q Q+QQQQQQ I+R S S + + P NL +N + LH P++
Sbjct: 190 QQLQKQQQQQQQAAAISRFPS-SIDAHLRPIRPINLQQNPNPNPNNPILNLH--QNPNS- 357
Query: 489 THLQSQSRHQMMQQNMNHLRSSYPSN 514
HLQ Q + Q QQ + P N
Sbjct: 358 NHLQQQQQQQQPQQQQQQQKMVRPGN 435
>BP033767
Length = 541
Score = 33.9 bits (76), Expect = 0.086
Identities = 25/55 (45%), Positives = 28/55 (50%)
Frame = +1
Query: 483 SGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAA 537
S PS PT S S + N N SS S +P SP PSSL SS+A AAA
Sbjct: 340 SPPSPPTSPPSSS---LKPPNPNPPTSSSASPLPPSPAPSPSSLSPPSSTADAAA 495
>CB829359
Length = 507
Score = 33.1 bits (74), Expect = 0.15
Identities = 25/73 (34%), Positives = 33/73 (44%), Gaps = 16/73 (21%)
Frame = -3
Query: 316 FFAHRPEELRPVYASTGSAMPS--PKSY-SSNAVDMSVMSPLALSSSSLP---------- 362
FF H+P+E+ + S PS P SY SS VD+S++ P L P
Sbjct: 271 FFKHKPDEMEQIVRLVWSYYPSTNPDSYPSSTNVDISILHPKFLIKKFYPQESRFNFYLT 92
Query: 363 ---MSTVSTPPMS 372
+S VS PP S
Sbjct: 91 TITLSLVSYPPNS 53
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 32.7 bits (73), Expect = 0.19
Identities = 18/49 (36%), Positives = 27/49 (54%)
Frame = +1
Query: 349 SVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPP 397
S S +LSSSS P S+ S+PP S + S S S + W + + + + P
Sbjct: 289 SSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSP 435
>TC8857 similar to UP|Q8LKG0 (Q8LKG0) Drm4, partial (84%)
Length = 772
Score = 32.3 bits (72), Expect = 0.25
Identities = 29/86 (33%), Positives = 39/86 (44%), Gaps = 9/86 (10%)
Frame = +1
Query: 307 ETGCG--RKVCFFAHRPEELRPVYASTGSAMP----SPKSYSSNAVDMSVMSPLALSS-- 358
E G G RK FA R + + + GS P SP+ + + ++ P S
Sbjct: 73 ENGLGKLRKHHTFASRSSPGKELADAAGSVRPYGVDSPEDATRVTRSIMIVKPPGYQSPS 252
Query: 359 SSLPMSTV-STPPMSPLAGSLSPKSG 383
S P S STPP+SP + LSP SG
Sbjct: 253 GSAPASPAGSTPPVSPFSAPLSPFSG 330
>TC18555 similar to GB|AAP68307.1|31711902|BT008868 At2g33570 {Arabidopsis
thaliana;}, partial (19%)
Length = 759
Score = 32.0 bits (71), Expect = 0.33
Identities = 22/65 (33%), Positives = 31/65 (46%)
Frame = +2
Query: 320 RPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLS 379
R E R Y+ + P + SS S SPL+ SSS L ++P P++ S S
Sbjct: 59 RKRERRSCYSLSSETTPPNSNSSSPPSSSSAPSPLSFSSSLL-----ASPSPLPISVSAS 223
Query: 380 PKSGN 384
P+S N
Sbjct: 224 PESPN 238
>AV415475
Length = 427
Score = 31.2 bits (69), Expect = 0.56
Identities = 18/30 (60%), Positives = 20/30 (66%)
Frame = +2
Query: 74 EKRTPLMIASLFGSTRVVKYLVETGRVDVN 103
+KRTPL +ASL G V K LVE G DVN
Sbjct: 179 DKRTPLHVASLHGWLAVAKCLVEFG-ADVN 265
>AV421966
Length = 469
Score = 31.2 bits (69), Expect = 0.56
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = -3
Query: 475 LLSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSL 526
LLS L L PT QS +H +MQ ++HLR + +SP P +L
Sbjct: 386 LLSYLPILGTGFEPTSRQSHRQHPLMQPGVHHLRELPLFQVEASP*PLPGTL 231
>BI417706
Length = 416
Score = 30.8 bits (68), Expect = 0.73
Identities = 24/90 (26%), Positives = 36/90 (39%)
Frame = +3
Query: 311 GRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPP 370
G + A EE+ A S+ S S + V+ S +P A S P TV+TP
Sbjct: 105 GAPIGLLAETEEEVAEAKAKAASSANSAASSAPPVVESSSPAPAAKVVSDGPRKTVATPY 284
Query: 371 MSPLAGSLSPKSGNLWQNKINLNLTPPSLQ 400
LA G++ N +TP ++
Sbjct: 285 AKKLAKQHKVDIGSVVGTGPNGRITPADVE 374
>TC15800 homologue to GB|AAM19932.1|20453387|AY097416 AT4g17800/dl4935c
{Arabidopsis thaliana;}, partial (39%)
Length = 818
Score = 30.8 bits (68), Expect = 0.73
Identities = 21/59 (35%), Positives = 26/59 (43%), Gaps = 15/59 (25%)
Frame = +1
Query: 407 KTALSARDFDLEMEMLGLGSPTR---------------QQQQQQQQQQLIEEIARISSP 450
K +S FD+ M L LG+P+ QQQQQQQ Q+ EE S P
Sbjct: 268 KNCISLSSFDMSMAGLDLGTPSSSHFLQNLHREDHFNLQQQQQQQPQESEEEPNHNSDP 444
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.129 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,011,055
Number of Sequences: 28460
Number of extensions: 188261
Number of successful extensions: 1376
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 1170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1291
length of query: 664
length of database: 4,897,600
effective HSP length: 96
effective length of query: 568
effective length of database: 2,165,440
effective search space: 1229969920
effective search space used: 1229969920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0204.2


