

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
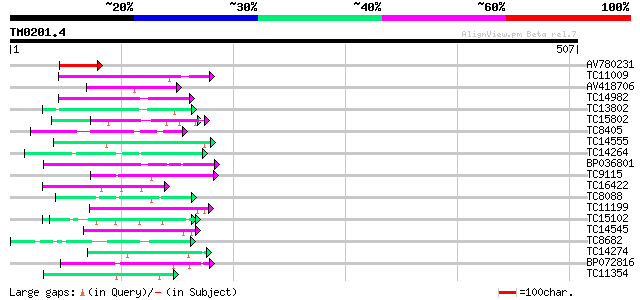
Query= TM0201.4
(507 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV780231 64 8e-11
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 52 3e-07
AV418706 50 9e-07
TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%) 48 3e-06
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 47 6e-06
TC15802 similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber... 47 1e-05
TC8405 weakly similar to UP|O04083 (O04083) Lysophospholipase is... 46 2e-05
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 45 2e-05
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 45 4e-05
BP036801 45 4e-05
TC9115 similar to PIR|T00425|T00425 photolyase/blue-light recept... 44 5e-05
TC16422 similar to PIR|T05653|T05653 amino acid transport protei... 44 8e-05
TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like arabinogalac... 43 1e-04
TC11199 42 2e-04
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 42 2e-04
TC14545 similar to UP|Q9FT78 (Q9FT78) P23 co-chaperone, partial ... 42 3e-04
TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate... 41 4e-04
TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.3... 41 5e-04
BP072816 41 5e-04
TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II ... 40 7e-04
>AV780231
Length = 530
Score = 63.5 bits (153), Expect = 8e-11
Identities = 31/39 (79%), Positives = 34/39 (86%)
Frame = -3
Query: 45 SNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPAS 83
SNPGAQPTTAAA KGKNV E+SVAAA EPT VPAS+P +
Sbjct: 477 SNPGAQPTTAAARKGKNVNEASVAAAIEPTTVPASTPVT 361
Score = 26.9 bits (58), Expect = 7.9
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Frame = -3
Query: 63 AESSVAAATEPTAV-PASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPI 118
A+SS A AV PAS+P + TAA ++ + +A++ T AS TP+
Sbjct: 528 AKSSFARKYFRVAVHPASNPGAQPTTAAARKGKNVNEASVAAAIEPTTVPAS--TPV 364
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 51.6 bits (122), Expect = 3e-07
Identities = 36/143 (25%), Positives = 69/143 (48%), Gaps = 3/143 (2%)
Frame = -2
Query: 44 TSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASA-GATAAVAAESSIGATAAS 102
+S+P + +++ P ++ A P+++ SSP+S+ G+ + A+ SS AT
Sbjct: 432 SSSPPSSESSSPEPSLSDLFLFDGTPAFSPSSLSLSSPSSSSGSKSLDASSSSAAATVRR 253
Query: 103 ASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPP--STHDAGSMPSPPHQGEKSC 160
+ K+++SS + + + SPP P +PP S+ + S SPP
Sbjct: 252 TRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPP------- 94
Query: 161 PGAATTSEAAQIEQAPAPEVGSS 183
PGA+++S ++ +P+ SS
Sbjct: 93 PGASSSSSSSPSSSSPSSSSSSS 25
>AV418706
Length = 374
Score = 50.1 bits (118), Expect = 9e-07
Identities = 28/89 (31%), Positives = 43/89 (47%), Gaps = 4/89 (4%)
Frame = +1
Query: 69 AATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKA----AASSDTPIGDKEKE 124
A T P VP P +A A AA SS G++++S+S +++ A +++S T +
Sbjct: 94 APTAPPTVPVLHPLAAAAATVAAAHSSAGSSSSSSSSSSSSAPPAPSSTSSTTLNALPSP 273
Query: 125 NETPKSPPRQDAPPSPPSTHDAGSMPSPP 153
+ PP PP PPS + S PP
Sbjct: 274 SRHSNFPPSNSPPPPPPSPPSSNSHSPPP 360
>TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%)
Length = 1036
Score = 48.1 bits (113), Expect = 3e-06
Identities = 33/123 (26%), Positives = 57/123 (45%), Gaps = 1/123 (0%)
Frame = +3
Query: 44 TSNPGAQPTTAAAPKGKNVAESSVAAATEPTAV-PASSPASAGATAAVAAESSIGATAAS 102
T P P A P ++ +T PTA+ A +P++A AA +A S+ G +++S
Sbjct: 45 TLPPKPPPPQTAPPPPTQLSPPQRPNSTAPTALLTALNPSTAAPAAAASAPSASG*SSSS 224
Query: 103 ASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPG 162
+S +++ A +S + + P +PP P + P++ PSPP+ S P
Sbjct: 225 SSSSSSSAVLASPST------SSTAPTTPPSPSPPSNSPTSTSPPPQPSPPNSTSPSPPQ 386
Query: 163 AAT 165
T
Sbjct: 387 TPT 395
Score = 30.4 bits (67), Expect = 0.71
Identities = 21/55 (38%), Positives = 23/55 (41%), Gaps = 4/55 (7%)
Frame = +3
Query: 128 PKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATT----SEAAQIEQAPAP 178
PK PP Q APP P SPP + + P A T S AA A AP
Sbjct: 54 PKPPPPQTAPPPPTQL-------SPPQRPNSTAPTALLTALNPSTAAPAAAASAP 197
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 47.4 bits (111), Expect = 6e-06
Identities = 40/148 (27%), Positives = 58/148 (39%), Gaps = 10/148 (6%)
Frame = +2
Query: 30 PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAA 89
P LTK S +SNP +T ++P S A + P+SSPASA ++
Sbjct: 59 PLHSTLTKPPSQI-SSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASS 235
Query: 90 VAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDA--- 146
+ +ASA ++T S+ T E+ + A P+PP++ A
Sbjct: 236 PTKSPKPSSPSASAPPSSTPLTPSTAT------SESSPTATSSSSSASPAPPTSFSALSR 397
Query: 147 -------GSMPSPPHQGEKSCPGAATTS 167
S PSPP S P A S
Sbjct: 398 ALELKALASSPSPPSSPPPSPPSAICMS 481
>TC15802 similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber hypocotyls
(Arabinogalactan protein), partial (26%)
Length = 795
Score = 46.6 bits (109), Expect = 1e-05
Identities = 40/138 (28%), Positives = 55/138 (38%), Gaps = 3/138 (2%)
Frame = +2
Query: 38 ASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIG 97
AS + P A P+TA AP + + P AVP +SP A + S
Sbjct: 188 ASPKPAATTPAASPSTATAPAPATTTPVTPVTSPPPAAVPVASPPPAAVPVSSPPAKSPP 367
Query: 98 ATAASA-SVNATKAAASSDTPIGDKEKENETP-KSPPRQDAPPS-PPSTHDAGSMPSPPH 154
A A +A V A P K K++ P SP D+PP+ P DA S P P
Sbjct: 368 APAPTAVPVAAPVTTPEVPAPAPSKTKKDAAPAPSPVVPDSPPAGAPGPSDAVS-PGPDG 544
Query: 155 QGEKSCPGAATTSEAAQI 172
G + A T + ++
Sbjct: 545 AGTANDESGAETMRSLKM 598
Score = 42.4 bits (98), Expect = 2e-04
Identities = 41/120 (34%), Positives = 52/120 (43%), Gaps = 14/120 (11%)
Frame = +2
Query: 73 PTAVPASSPASAGAT-------AAVAAESSIGATAASASVNATKAAASSDTPIGDKEKEN 125
PTA P +SPA+ AT A VA+ T A++ AT A ++ TP+
Sbjct: 113 PTAAPTTSPATPSATTPVSTPVAPVASPKPAATTPAASPSTATAPAPATTTPV------- 271
Query: 126 ETPKSPPRQDAPP---SPPSTHDAGSMP--SPPHQGEKSCPGAA--TTSEAAQIEQAPAP 178
TP + P A P PP+ S P SPP + P AA TT E APAP
Sbjct: 272 -TPVTSPPPAAVPVASPPPAAVPVSSPPAKSPPAPAPTAVPVAAPVTTPEV----PAPAP 436
Score = 37.4 bits (85), Expect = 0.006
Identities = 36/144 (25%), Positives = 54/144 (37%), Gaps = 8/144 (5%)
Frame = +2
Query: 43 GTSNPGAQPTTA-AAPKGKNVAESSVA-------AATEPTAVPASSPASAGATAAVAAES 94
G +P A PTT+ A P + VA AAT P A P+++ A A AT
Sbjct: 101 GGQSPTAAPTTSPATPSATTPVSTPVAPVASPKPAATTPAASPSTATAPAPATTTPVTPV 280
Query: 95 SIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPH 154
+ AA + AA +P ++P +P P + P T P+P
Sbjct: 281 TSPPPAAVPVASPPPAAVPVSSPPA------KSPPAPAPTAVPVAAPVTTPEVPAPAPSK 442
Query: 155 QGEKSCPGAATTSEAAQIEQAPAP 178
+ + P + + AP P
Sbjct: 443 TKKDAAPAPSPVVPDSPPAGAPGP 514
Score = 33.9 bits (76), Expect = 0.064
Identities = 25/94 (26%), Positives = 39/94 (40%), Gaps = 3/94 (3%)
Frame = +2
Query: 88 AAVAAESSIGA-TAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDA 146
A V +S A T + A+ +AT ++ P+ + TP + P P+P +T
Sbjct: 92 AGVGGQSPTAAPTTSPATPSATTPVSTPVAPVASPKPAATTPAASPSTATAPAPATTTPV 271
Query: 147 GSMPSPPHQG--EKSCPGAATTSEAAQIEQAPAP 178
+ SPP S P AA + + PAP
Sbjct: 272 TPVTSPPPAAVPVASPPPAAVPVSSPPAKSPPAP 373
>TC8405 weakly similar to UP|O04083 (O04083) Lysophospholipase isolog,
25331-24357 (At1g11090), partial (18%)
Length = 552
Score = 45.8 bits (107), Expect = 2e-05
Identities = 38/142 (26%), Positives = 60/142 (41%), Gaps = 1/142 (0%)
Frame = +3
Query: 19 GAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVP- 77
GA + +T PK +S T+ P++ + PK K P++ P
Sbjct: 75 GATCRKRNTTPPKESATPNPTSTLPTARSSPSPSSPSTPKSK-----------PPSS*PT 221
Query: 78 ASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAP 137
A++P AG++ A+ S GAT +S ++ AA P + SPP P
Sbjct: 222 ATAPTPAGSSRRSASTSPPGATPSSPPTSSATAA-----PTASAATSATSTASPP----P 374
Query: 138 PSPPSTHDAGSMPSPPHQGEKS 159
SP S+ A + P+PP + S
Sbjct: 375 RSPSSSTSAAASPTPPFRPSSS 440
Score = 33.9 bits (76), Expect = 0.064
Identities = 27/110 (24%), Positives = 42/110 (37%), Gaps = 6/110 (5%)
Frame = +3
Query: 81 PASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSP 140
P ++GAT + +A + A SS +P +++ P S P AP
Sbjct: 63 PLTSGATCRKRNTTPPKESATPNPTSTLPTARSSPSPSSPSTPKSKPPSS*PTATAPTPA 242
Query: 141 ------PSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 184
ST G+ PS P + A+ + +A +P P SSS
Sbjct: 243 GSSRRSASTSPPGATPSSPPTSSATAAPTASAATSATSTASPPPRSPSSS 392
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 45.4 bits (106), Expect = 2e-05
Identities = 38/155 (24%), Positives = 61/155 (38%), Gaps = 10/155 (6%)
Frame = +2
Query: 40 SDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAG---ATAAVAAESSI 96
+ G +P PTT+ AP + A ++ P A + P + A+ A+SS
Sbjct: 170 ASVGAQSPSNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQSSP 349
Query: 97 GATAASASVNATKAAASSDTPI-GDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSP-PH 154
++ V +T A + TP + P +PP A P P T + P+P P
Sbjct: 350 PPVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPALTPVPATSPAPAPA 529
Query: 155 QGEKSCPGAATTSEAAQI-----EQAPAPEVGSSS 184
+ + P A A + +AP P + S S
Sbjct: 530 KVKSKSPAPAPAPALAPVVSLSPSEAPGPSLSSLS 634
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 44.7 bits (104), Expect = 4e-05
Identities = 46/165 (27%), Positives = 66/165 (39%), Gaps = 1/165 (0%)
Frame = +2
Query: 14 AKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEP 73
A K+ A+S PK + A++ + P + AA K A++ AAA +P
Sbjct: 485 AAKKKPQPAAAKSKPKPKAKAKPAAAATKAKAKPAPKSKAAAT---KTTAKAKPAAAAKP 655
Query: 74 TAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPR 133
A PA+ A AVAA + A AS KA A S T PK+ P
Sbjct: 656 KAKPAAKAKPAAKAKAVAAPAK-----AKASPAKPKAKAKSKT-APRMNVNPPVPKAKPA 817
Query: 134 QDAPPSP-PSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPA 177
A P+ P+ S + P + P A A +++APA
Sbjct: 818 AKAKPAARPAKASRTSTRTTPGKKVPPPPKPAPKKAATPVKKAPA 952
>BP036801
Length = 540
Score = 44.7 bits (104), Expect = 4e-05
Identities = 46/158 (29%), Positives = 68/158 (42%), Gaps = 1/158 (0%)
Frame = +2
Query: 31 KRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAV 90
K + L +SS S A P AAA G + ++ A+ T ++ + +TA
Sbjct: 8 KNKPLLLSSSP*SPSCCSASPQHAAAQGGTGFSYNAA*ASQPCTCRS*TTTRWSSSTAPT 187
Query: 91 AAE-SSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSM 149
+A +S TAA+AS T ++S+ P + T P PS P TH A
Sbjct: 188 SAHPTSPSPTAAAASTPVTWPSSSTAPPT-----PSSTTSPPTLSALSPSKP-THGAPPA 349
Query: 150 PSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYN 187
PSPP S P A+TT+ + PA S++ N
Sbjct: 350 PSPP-TAPSSKPAASTTATTSSAPSPPAHNTTSATGSN 460
Score = 37.4 bits (85), Expect = 0.006
Identities = 35/130 (26%), Positives = 58/130 (43%), Gaps = 7/130 (5%)
Frame = +2
Query: 26 STKAPKRRRLTKAS-SDAGTSNPGAQPTTAAAPKGKNVAES----SVAAATEPT--AVPA 78
S+ AP T S + A S P P+++ AP + S S + ++PT A PA
Sbjct: 170 SSTAPTSAHPTSPSPTAAAASTPVTWPSSSTAPPTPSSTTSPPTLSALSPSKPTHGAPPA 349
Query: 79 SSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPP 138
SP +A ++ A+ ++ ++A S + T +A S++P SPP PP
Sbjct: 350 PSPPTAPSSKPAASTTATTSSAPSPPAHNTTSATGSNSP----------RISPPLAGTPP 499
Query: 139 SPPSTHDAGS 148
+ S + S
Sbjct: 500 TKSSPTEESS 529
>TC9115 similar to PIR|T00425|T00425 photolyase/blue-light receptor (PHR2)
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (55%)
Length = 1018
Score = 44.3 bits (103), Expect = 5e-05
Identities = 30/116 (25%), Positives = 48/116 (40%), Gaps = 2/116 (1%)
Frame = +1
Query: 73 PTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKEN--ETPKS 130
P P SSP+S + + ++ T + + + S P + N P S
Sbjct: 169 PFHSPPSSPSSNPNSHHLLPHNN-NPTRSKSQPKPPPSPTSHSPPPPHPRRPNPPSNPPS 345
Query: 131 PPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYY 186
PP AP +P + ++P+PPH G+ATTS +A + P S +Y
Sbjct: 346 PPTPSAPLTP*APTAPSTLPTPPHSAAPPSSGSATTSASATTRLSTPPTTNRSQFY 513
Score = 30.8 bits (68), Expect = 0.54
Identities = 21/70 (30%), Positives = 27/70 (38%), Gaps = 12/70 (17%)
Frame = -2
Query: 117 PIGDKEKENETPKSPPRQDAPP-SPPSTHDAGSMPSPPHQGEKSC-----------PGAA 164
P +K K P+ +PP SPPS+ S P PH + C P
Sbjct: 861 PPTEKANPQHGTKCSPKSTSPPHSPPSSPPQSSPPPSPHHAKPLCEHTQRQHQQPSPAPP 682
Query: 165 TTSEAAQIEQ 174
T S A Q+ Q
Sbjct: 681 TQSPACQL*Q 652
>TC16422 similar to PIR|T05653|T05653 amino acid transport protein homolog
F22I13.20 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (38%)
Length = 685
Score = 43.5 bits (101), Expect = 8e-05
Identities = 37/136 (27%), Positives = 64/136 (46%), Gaps = 22/136 (16%)
Frame = +3
Query: 30 PKRRRLTKASSDAGTSNPGAQPT-TAAAPKGKNVAESSVAAATEPTAVPASS-------- 80
P + +S + S P ++P+ T+++P + +S+ A++EP PASS
Sbjct: 135 PTLEKTHLSSPNPHLSPPNSKPSPTSSSPSSAPESSASLTASSEPAGSPASSCSSPSPSS 314
Query: 81 PASAGATAAVAAESSIGA--------TAASASVNATKAAASSDT-----PIGDKEKENET 127
P +A +++ A SSI + +A SAS +A ++AAS T P D +
Sbjct: 315 PTTA*CSSSPLAASSIPSLGTPNSDPSATSASPSAARSAASPSTP*LSSPRLDSASATSS 494
Query: 128 PKSPPRQDAPPSPPST 143
P +P +PP+T
Sbjct: 495 SSRPLSASSPATPPAT 542
>TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (69%)
Length = 1527
Score = 42.7 bits (99), Expect = 1e-04
Identities = 36/129 (27%), Positives = 52/129 (39%), Gaps = 3/129 (2%)
Frame = +2
Query: 42 AGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAA 101
A T+ P P T +P + S + A TA P SP+S T S+ +T+
Sbjct: 140 AATTLPAFSPNTPTSPPSTTTSPSPTSPARSTTAKP--SPSSPSTTPPCPPSST--STSP 307
Query: 102 SASVNATKAAASSDTPIGDKEKEN---ETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEK 158
S + + SS T + K E P SPP PP+PP A S + +
Sbjct: 308 SPP*KTSSPSTSSSTTLAPKSSTR*PTEPPSSPPCS-RPPAPPPEPPATSTSPTSKEAK* 484
Query: 159 SCPGAATTS 167
+ P TT+
Sbjct: 485 ASPLRTTTA 511
Score = 29.3 bits (64), Expect = 1.6
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 2/60 (3%)
Frame = +2
Query: 127 TPKSPPRQDAPPSPPS-THDAGSMPSPPHQG-EKSCPGAATTSEAAQIEQAPAPEVGSSS 184
TP SPP PSP S + PSP CP ++T++ + + +P SS+
Sbjct: 173 TPTSPPSTTTSPSPTSPARSTTAKPSPSSPSTTPPCPPSSTSTSPSPP*KTSSPSTSSST 352
>TC11199
Length = 743
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/118 (30%), Positives = 49/118 (41%), Gaps = 7/118 (5%)
Frame = +1
Query: 72 EPTAVPASSPASAGATAAVAAESSIGATAASA-SVNATKAAASSDTPIGDKEKENETPKS 130
+P++ SSPAS + +A S + S S T +SS + I D P S
Sbjct: 157 DPSSARPSSPASQSHSTTTSATSLAPKSPRSKRSARRTTRTSSSPSTISDPSSPTSNPSS 336
Query: 131 PPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATT----SEAAQI--EQAPAPEVGS 182
PP PS + S PS P PG +T+ SE+A + APAP S
Sbjct: 337 PPSLTPTPSSSPSLALSSPPSTPSSKRGMYPGTSTSPSTRSESAFS*WKIAPAPTATS 510
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 42.4 bits (98), Expect = 2e-04
Identities = 42/146 (28%), Positives = 59/146 (39%), Gaps = 11/146 (7%)
Frame = +1
Query: 36 TKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAE-- 93
T SS + T P +T PK KN AT TA PAS ATA ++
Sbjct: 295 TPKSSHSSTPPPSRSSSTT--PKSKN--------ATSLTAPSCPPPASPAATAPAPSKTP 444
Query: 94 -----SSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQD----APPSPPSTH 144
+S ++ +S+ T ++ S P TP SP Q+ PPS P
Sbjct: 445 PNPPTNSSKPSSCKSSLPNTTPSSLSSAPPSPTTSPISTPSSPENQEKPASTPPSAPPRS 624
Query: 145 DAGSMPSPPHQGEKSCPGAATTSEAA 170
+ S SP + K P +AT + A+
Sbjct: 625 TSSSSSSPINTPRK--PSSATKAPAS 696
Score = 40.4 bits (93), Expect = 7e-04
Identities = 37/144 (25%), Positives = 57/144 (38%), Gaps = 6/144 (4%)
Frame = +1
Query: 30 PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAV--PASSPASAGAT 87
P SS+ T+ T AA + S+ ++EPT + PAS P +
Sbjct: 133 PSLEATASPSSEPCTTATTTSTTKAATNSSPPESRSTTPPSSEPTCLLAPASPPTPKSSH 312
Query: 88 AAVAAESSIGATAASASVNATKAAASS----DTPIGDKEKENETPKSPPRQDAPPSPPST 143
++ S +T S NAT A S +P ++TP +PP + PS +
Sbjct: 313 SSTPPPSRSSSTTPK-SKNATSLTAPSCPPPASPAATAPAPSKTPPNPPTNSSKPSSCKS 489
Query: 144 HDAGSMPSPPHQGEKSCPGAATTS 167
S+P+ S P + TTS
Sbjct: 490 ----SLPNTTPSSLSSAPPSPTTS 549
Score = 37.0 bits (84), Expect = 0.008
Identities = 37/148 (25%), Positives = 57/148 (38%)
Frame = +1
Query: 44 TSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASA 103
T++P ++P T A S+ AAT SSP + +T ++E + A A
Sbjct: 148 TASPSSEPCTTAT------TTSTTKAATN------SSPPESRSTTPPSSEPT--CLLAPA 285
Query: 104 SVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGA 163
S K++ SS P K+ AP PP A + P+P K+ P
Sbjct: 286 SPPTPKSSHSSTPPPSRSSSTTPKSKNATSLTAPSCPPPASPAATAPAP----SKTPPNP 453
Query: 164 ATTSEAAQIEQAPAPEVGSSSYYNMLPN 191
T S ++ P SS + P+
Sbjct: 454 PTNSSKPSSCKSSLPNTTPSSLSSAPPS 537
Score = 37.0 bits (84), Expect = 0.008
Identities = 40/136 (29%), Positives = 59/136 (42%), Gaps = 18/136 (13%)
Frame = +1
Query: 79 SSPASAGAT-------AAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKS- 130
++P +A AT A A+ SS T A+ + + TKAA +S P E + TP S
Sbjct: 94 TTPKTAAATFL*NPSLEATASPSSEPCTTATTT-STTKAATNSSPP----ESRSTTPPSS 258
Query: 131 -PPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSE---------AAQIEQAPAPEV 180
P AP SPP+ + S PP + + P + + A+ APAP
Sbjct: 259 EPTCLLAPASPPTPKSSHSSTPPPSRSSSTTPKSKNATSLTAPSCPPPASPAATAPAP-- 432
Query: 181 GSSSYYNMLPNAIEPS 196
S + N N+ +PS
Sbjct: 433 -SKTPPNPPTNSSKPS 477
>TC14545 similar to UP|Q9FT78 (Q9FT78) P23 co-chaperone, partial (46%)
Length = 1127
Score = 41.6 bits (96), Expect = 3e-04
Identities = 32/114 (28%), Positives = 51/114 (44%), Gaps = 10/114 (8%)
Frame = -2
Query: 67 VAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENE 126
+ AA+ + V + + +A +A + A + +S+S+ + + ASS P ++
Sbjct: 781 LGAASAFSVVASLALLTASCSAGLDASPGFDTSCSSSSLPSKSSIASSPIPPNLEKSIPP 602
Query: 127 TPKSPPRQDAPPSPPSTHDAGSMPSPPH-------QGEKSCP---GAATTSEAA 170
P PP PP PP S P PP EKS P G+ATTS ++
Sbjct: 601 NPSIPPNPPIPPIPPIPPIPPSPPRPPSPPIPIPPNFEKSIPPISGSATTSSSS 440
>TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate
dioxygenase (4HPPD) (HPD) (HPPDase) , partial (29%)
Length = 576
Score = 41.2 bits (95), Expect = 4e-04
Identities = 44/166 (26%), Positives = 61/166 (36%)
Frame = +1
Query: 1 MARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGK 60
+ + P N+ G G +NQ T +P + T +S G P PT AA G
Sbjct: 163 LKKSKPGSNSSGSTASSGPTQNQ---TGSP*KGSTTSSS---GAPTP---PTPHAASPGA 315
Query: 61 NVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGD 120
+ SS + P T SS AT+AS+S T + S P
Sbjct: 316 SACRSSQNQTSPPE------------TTLTPPTSSAPATSASSSPPHTLPKSPSPPP--- 450
Query: 121 KEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATT 166
+P SPP +P PP+ + PS K P +TT
Sbjct: 451 ----PPSPLSPPPPASPSPPPTASPSAPSPSKSPTPSKPSPPQSTT 576
Score = 36.2 bits (82), Expect = 0.013
Identities = 30/116 (25%), Positives = 45/116 (37%), Gaps = 2/116 (1%)
Frame = +1
Query: 65 SSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKE 124
S A++ PT SP T++ A + AAS +A +++ + +P +
Sbjct: 193 SGSTASSGPTQNQTGSP*KGSTTSSSGAPTPPTPHAASPGASACRSSQNQTSP----PET 360
Query: 125 NETP--KSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 178
TP S P A SPP T P PP P A+ + AP+P
Sbjct: 361 TLTPPTSSAPATSASSSPPHTLPKSPSPPPPPSPLSPPPPASPSPPPTASPSAPSP 528
>TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.35
{Arabidopsis thaliana;} , partial (16%)
Length = 1208
Score = 40.8 bits (94), Expect = 5e-04
Identities = 33/133 (24%), Positives = 52/133 (38%), Gaps = 22/133 (16%)
Frame = +3
Query: 70 ATEPTAVPASSPASAGATAAVAAESSIGATAASAS----------------VNATKAAAS 113
+T P++ P+ S S G A + SS G+T + S V+ + +
Sbjct: 78 STSPSSPPSISRTSTGKPATSSRTSSSGSTPTAVSPPNPTTPATPPPSGTSVSPSLSLTP 257
Query: 114 SDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEK------SCPGAATTS 167
S TP P PP +PPS S+ + + PP + P A +TS
Sbjct: 258 SLTPFSPSRSSTPNPPIPPNPSSPPSASSSPTSTTSMIPPASANSPSSAPPAAPTARSTS 437
Query: 168 EAAQIEQAPAPEV 180
+A + AP P +
Sbjct: 438 SSASL-VAPNPSI 473
>BP072816
Length = 492
Score = 40.8 bits (94), Expect = 5e-04
Identities = 36/142 (25%), Positives = 58/142 (40%), Gaps = 4/142 (2%)
Frame = +1
Query: 46 NPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASV 105
NP +P + + S + + P++SP+SA + A S T A+
Sbjct: 82 NPPLSHQPCHSPSSSSSSSSPLPSPPTTLHAPSTSPSSAPSATAAPPPS----TPTPAAN 249
Query: 106 NATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHD--AGSMPSPPHQGEKS--CP 161
++K + SS + +PP+ PPS PS+ + A + S PH K+ P
Sbjct: 250 TSSKPSTSSKPTTSAAPPASSLLSTPPKPAGPPSNPSSTNSPAPASTSAPHAASKAHGSP 429
Query: 162 GAATTSEAAQIEQAPAPEVGSS 183
A TS A P+P+ S
Sbjct: 430 KVA*TSPACN-NSKPSPQTPHS 492
Score = 33.9 bits (76), Expect = 0.064
Identities = 26/94 (27%), Positives = 44/94 (46%), Gaps = 2/94 (2%)
Frame = +1
Query: 26 STKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAG 85
+T AP T A++ + + ++PTT+AAP SS+ + A P S+P+S
Sbjct: 205 ATAAPPPSTPTPAANTSSKPSTSSKPTTSAAPPA-----SSLLSTPPKPAGPPSNPSSTN 369
Query: 86 ATAAVAAESSIGATAA--SASVNATKAAASSDTP 117
+ A + + A+ A S V T A ++ P
Sbjct: 370 SPAPASTSAPHAASKAHGSPKVA*TSPACNNSKP 471
>TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II , partial
(6%)
Length = 583
Score = 40.4 bits (93), Expect = 7e-04
Identities = 34/145 (23%), Positives = 56/145 (38%), Gaps = 24/145 (16%)
Frame = -1
Query: 31 KRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAV 90
KR +SS A S+PG + ++ A + + +E A P + P SP+ +
Sbjct: 469 KRCVSRSSSSSAPPSSPGRKKSSETAIRPHSASEPDPAPPPPPPSSPPMSPSPTATSPPT 290
Query: 91 AAES--------------SIGATAASASVNATKAAASSDTPIGDKEKENETPKSPP---- 132
++ + S T AS++ +TK S+ + + +P SPP
Sbjct: 289 SSPNTHPPTPTPNP*SSPSPSTTTASSASPSTKTLPSTRPRPASESPTSSSPSSPPPSSG 110
Query: 133 ------RQDAPPSPPSTHDAGSMPS 151
R PP PPS + PS
Sbjct: 109 SSVSPPRISPPPPPPSNSPTATPPS 35
Score = 32.7 bits (73), Expect = 0.14
Identities = 29/113 (25%), Positives = 49/113 (42%), Gaps = 2/113 (1%)
Frame = -1
Query: 86 ATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSP-PRQDAPP-SPPST 143
++++ A SS G +S + +A+ D P + P SP P +PP S P+T
Sbjct: 451 SSSSSAPPSSPGRKKSSETAIRPHSASEPD-PAPPPPPPSSPPMSPSPTATSPPTSSPNT 275
Query: 144 HDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 196
H P+P S A++ S + + + P S S + P++ PS
Sbjct: 274 HPPTPTPNP*SSPSPSTTTASSASPSTKTLPSTRPRPASESPTSSSPSSPPPS 116
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.304 0.121 0.318
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,443,109
Number of Sequences: 28460
Number of extensions: 97276
Number of successful extensions: 2556
Number of sequences better than 10.0: 395
Number of HSP's better than 10.0 without gapping: 1525
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2080
length of query: 507
length of database: 4,897,600
effective HSP length: 94
effective length of query: 413
effective length of database: 2,222,360
effective search space: 917834680
effective search space used: 917834680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0201.4


