

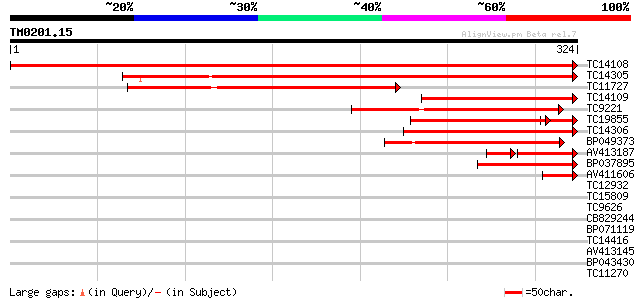
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.15
(324 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14108 similar to UP|CAHC_PEA (P17067) Carbonic anhydrase, chlo... 642 0.0
TC14305 homologue to UP|Q84VT6 (Q84VT6) Carbonic anhydrase , co... 276 3e-75
TC11727 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , pa... 215 7e-57
TC14109 similar to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partia... 185 8e-48
TC9221 similar to UP|O81875 (O81875) Carbonate dehydratase - lik... 119 5e-28
TC19855 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , pa... 104 1e-25
TC14306 homologue to UP|Q84VT6 (Q84VT6) Carbonic anhydrase , pa... 93 5e-20
BP049373 92 2e-19
AV413187 72 8e-19
BP037895 55 1e-08
AV411606 43 6e-05
TC12932 36 0.008
TC15809 weakly similar to PIR|JN0747|JN0747 histone H1-I - Volvo... 35 0.022
TC9626 weakly similar to UP|Q8LEE9 (Q8LEE9) Arabinogalactan prot... 32 0.19
CB829244 32 0.19
BP071119 31 0.32
TC14416 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, parti... 29 1.2
AV413145 29 1.2
BP043430 29 1.2
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 29 1.2
>TC14108 similar to UP|CAHC_PEA (P17067) Carbonic anhydrase, chloroplast
precursor (Carbonate dehydratase) , partial (98%)
Length = 1458
Score = 642 bits (1656), Expect = 0.0
Identities = 324/324 (100%), Positives = 324/324 (100%)
Frame = +1
Query: 1 MSTSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAP 60
MSTSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAP
Sbjct: 64 MSTSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAP 243
Query: 61 IIPTLDMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRI 120
IIPTLDMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRI
Sbjct: 244 IIPTLDMGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRI 423
Query: 121 KTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVA 180
KTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVA
Sbjct: 424 KTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVA 603
Query: 181 NMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIED 240
NMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIED
Sbjct: 604 NMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIED 783
Query: 241 WVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGG 300
WVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGG
Sbjct: 784 WVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGG 963
Query: 301 YYDFVKGSFELWSLQFDLASSFSV 324
YYDFVKGSFELWSLQFDLASSFSV
Sbjct: 964 YYDFVKGSFELWSLQFDLASSFSV 1035
>TC14305 homologue to UP|Q84VT6 (Q84VT6) Carbonic anhydrase , complete
Length = 1083
Score = 276 bits (706), Expect = 3e-75
Identities = 144/266 (54%), Positives = 181/266 (67%), Gaps = 6/266 (2%)
Frame = +3
Query: 65 LDMGKNYEE------AIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASD 118
L GKN + AIEEL+KL+REK EL +A K+E + A+L P + A
Sbjct: 66 LKKGKNMAKHSPDQLAIEELKKLIREKEELNESADTKIEDVIAELQLQGCHPTPET-AEQ 242
Query: 119 RIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRN 178
RI GF YFK +DKNP LY ELAKGQSP +++FACSDSRV P+ +L+FQPGEAF+VRN
Sbjct: 243 RIIDGFTYFKIHNFDKNPDLYKELAKGQSPKFLIFACSDSRVSPTTILNFQPGEAFMVRN 422
Query: 179 VANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFI 238
+ANMVPP++Q ++SG GAAIEYA+L LKV NI++IGHS CGGI L+S P D + S +FI
Sbjct: 423 IANMVPPFNQLRYSGVGAAIEYAILELKVPNILIIGHSRCGGIARLMSHPEDDSPSFEFI 602
Query: 239 EDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALK 298
+DWV IGLPAK KV +H E T CEKE+VN SL NL +YPFV + +K L L
Sbjct: 603 DDWVKIGLPAKIKVLKEHACCDSLEQRTLCEKESVNNSLVNLHTYPFVDRAIRSKGLGLF 782
Query: 299 GGYYDFVKGSFELWSLQFDLASSFSV 324
GGYYDFV G F+LW + + S+
Sbjct: 783 GGYYDFVNGEFKLWKYESHVTKHISI 860
>TC11727 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , partial (48%)
Length = 584
Score = 215 bits (548), Expect = 7e-57
Identities = 104/156 (66%), Positives = 127/156 (80%)
Frame = +2
Query: 68 GKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYF 127
G++Y+EAI L KLL EK +L A AA K++ +TA+L S E RI+TGF+ F
Sbjct: 119 GESYDEAIASLTKLLSEKADLGAVAAAKIKDLTAELEDAGSKPFNPDE---RIRTGFVQF 289
Query: 128 KKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYD 187
K EK++KNP LYGELAKGQSP ++VFACSDSRVCPSH+L+FQPGEAF+VRN+ANMVPP+D
Sbjct: 290 KSEKFEKNPDLYGELAKGQSPKFLVFACSDSRVCPSHILNFQPGEAFMVRNIANMVPPFD 469
Query: 188 QAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKG 223
+ K+SG+GAAIEYAVLHLKV NIVVIGHS CGGI+G
Sbjct: 470 KTKYSGAGAAIEYAVLHLKVENIVVIGHSCCGGIRG 577
>TC14109 similar to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (27%)
Length = 570
Score = 185 bits (470), Expect = 8e-48
Identities = 89/89 (100%), Positives = 89/89 (100%)
Frame = +1
Query: 236 DFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTL 295
DFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTL
Sbjct: 1 DFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTL 180
Query: 296 ALKGGYYDFVKGSFELWSLQFDLASSFSV 324
ALKGGYYDFVKGSFELWSLQFDLASSFSV
Sbjct: 181 ALKGGYYDFVKGSFELWSLQFDLASSFSV 267
>TC9221 similar to UP|O81875 (O81875) Carbonate dehydratase - like protein
(Carbonate dehydratase-like protein), partial (68%)
Length = 571
Score = 119 bits (299), Expect = 5e-28
Identities = 58/121 (47%), Positives = 83/121 (67%)
Frame = +1
Query: 196 AAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAK 255
AA+E+AV L++ NI VIGHS CGGI+ L+S D + S FI++WV IG A+ K +A
Sbjct: 1 AALEFAVNTLQLENIFVIGHSCCGGIRALMSMQDDASAS--FIKNWVVIGKNARIKTEAA 174
Query: 256 HGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQ 315
+ F E C+HCEKE+++ SL NLL+YP++ + L N L + GGYY+F+ SFE W+L
Sbjct: 175 ASNLSFDEQCSHCEKESIHHSLLNLLTYPWIEQKLANGELMIHGGYYNFIDCSFEKWTLD 354
Query: 316 F 316
+
Sbjct: 355 Y 357
>TC19855 similar to UP|Q84Y10 (Q84Y10) Carbonic anhydrase 2 , partial (27%)
Length = 642
Score = 104 bits (259), Expect(2) = 1e-25
Identities = 50/80 (62%), Positives = 60/80 (74%)
Frame = +1
Query: 230 DGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREG 289
DGTT++DFIE WV I PAK+KVK G F E CT+CEKEAVNVSLGNLL+YPFV+E
Sbjct: 4 DGTTASDFIEQWVQICNPAKSKVKTDTGSLSFSEQCTNCEKEAVNVSLGNLLTYPFVKER 183
Query: 290 LVNKTLALKGGYYDFVKGSF 309
+++K LALKG +Y F F
Sbjct: 184 VLDKPLALKGAHYPFCYWQF 243
Score = 28.5 bits (62), Expect(2) = 1e-25
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 304 FVKGSFELWSLQFDLASSFSV 324
FV G+FELW L F + S SV
Sbjct: 227 FVTGNFELWDLNFKVQPSISV 289
>TC14306 homologue to UP|Q84VT6 (Q84VT6) Carbonic anhydrase , partial (39%)
Length = 622
Score = 93.2 bits (230), Expect = 5e-20
Identities = 48/99 (48%), Positives = 60/99 (60%)
Frame = +2
Query: 226 SFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPF 285
S P D + S +FI+DWV IGLPA+ KV +H E T CEKE+VN SL NL +YPF
Sbjct: 2 SHPEDDSPSFEFIDDWVKIGLPAQIKVLKEHACCDSLEQRTLCEKESVNNSLVNLHTYPF 181
Query: 286 VREGLVNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
V + +K L L GGYYDFV G F+LW + S+
Sbjct: 182 VDRAIRSKGLGLFGGYYDFVNGEFKLWKYESHFTQHISI 298
>BP049373
Length = 495
Score = 91.7 bits (226), Expect = 2e-19
Identities = 46/103 (44%), Positives = 63/103 (60%)
Frame = -1
Query: 215 HSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVN 274
HS+C GI+ L+ D S +FI W++ G AK K A F + C CEKE++N
Sbjct: 495 HSSCAGIENLMRMQED-VESRNFIFKWLANGKVAKLKTNAATAHLTFDQQCRFCEKESIN 319
Query: 275 VSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQFD 317
SL NLLSYP++++ + K L+L GGYYDF SFE W+L F+
Sbjct: 318 QSLLNLLSYPWIQDRVSKKLLSLHGGYYDFSNCSFEKWTLDFE 190
>AV413187
Length = 339
Score = 72.0 bits (175), Expect(2) = 8e-19
Identities = 34/34 (100%), Positives = 34/34 (100%)
Frame = -2
Query: 291 VNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
VNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV
Sbjct: 284 VNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 183
Score = 37.7 bits (86), Expect(2) = 8e-19
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = -1
Query: 273 VNVSLGNLLSYPFVREG 289
VNVSLGNLLSYPFVREG
Sbjct: 339 VNVSLGNLLSYPFVREG 289
>BP037895
Length = 251
Score = 55.5 bits (132), Expect = 1e-08
Identities = 35/58 (60%), Positives = 38/58 (65%), Gaps = 1/58 (1%)
Frame = -1
Query: 268 CEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKG-SFELWSLQFDLASSFSV 324
CEKEAVNVS GNLLSYP VREGLV++TLAL G + L FD A SFSV
Sbjct: 251 CEKEAVNVSPGNLLSYPIVREGLVDQTLAL*RRIL*LC*GITLSSGGLPFDPAFSFSV 78
>AV411606
Length = 320
Score = 43.1 bits (100), Expect = 6e-05
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = -1
Query: 305 VKGSFELWSLQFDLASSFSV 324
VKGSFELWSLQFDLASSFSV
Sbjct: 320 VKGSFELWSLQFDLASSFSV 261
>TC12932
Length = 451
Score = 36.2 bits (82), Expect = 0.008
Identities = 40/144 (27%), Positives = 54/144 (36%), Gaps = 7/144 (4%)
Frame = +1
Query: 10 YLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFP---SLIQDRPVFAAPAPIIPTLD 66
Y SS + + +TL AS + P SSSS S I R +
Sbjct: 55 YSSSFPNFTSLMASITLSSDPFASKTLPISSSSYCIQLGTSRISARALITG--------- 207
Query: 67 MGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEAS----DRIKT 122
E+ L LR A+ +T QL + D + +E + +K
Sbjct: 208 ---RIEQTHLRLSTELRRNQGFTLKASMGPPGLTEQLNNSKLDTLAEAEDECDIFNDLKD 378
Query: 123 GFLYFKKEKYDKNPALYGELAKGQ 146
FL FKK KY KNP + LAK Q
Sbjct: 379 RFLSFKKNKYMKNPEQFESLAKAQ 450
>TC15809 weakly similar to PIR|JN0747|JN0747 histone H1-I - Volvox carteri
{Volvox carteri;}, partial (10%)
Length = 526
Score = 34.7 bits (78), Expect = 0.022
Identities = 27/104 (25%), Positives = 47/104 (44%), Gaps = 12/104 (11%)
Frame = +2
Query: 34 ASSPSSSSSSSFPSLIQDRPVFAAPAP--IIPTLDMGKNYEEAI-EELQKLLREKNELKA 90
A SP+ +++ P+ P AP+P ++PTL + ++ +++ K+LR+
Sbjct: 152 AISPAPAAAPKSPAKTPPTPKATAPSPKPLVPTLPQSPDSSDSTPDDITKILRKAKIFSV 331
Query: 91 -----TAAEKVEQITAQLGTTSSDGI----PSSEASDRIKTGFL 125
E + I +QL T + GI P A +K GFL
Sbjct: 332 LIRLLKTTEIMNNINSQLITAKNGGITILAPDDSAFSHLKAGFL 463
>TC9626 weakly similar to UP|Q8LEE9 (Q8LEE9) Arabinogalactan protein-like,
partial (24%)
Length = 605
Score = 31.6 bits (70), Expect = 0.19
Identities = 32/117 (27%), Positives = 46/117 (38%), Gaps = 4/117 (3%)
Frame = +1
Query: 12 SSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKNY 71
S P+ SL +PLV + SPSS SS QD II L K++
Sbjct: 163 SPAKPSPASLAPAPAKPLVPSLPQSPSSDSSG------QD---------IIKILRKAKSF 297
Query: 72 EEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGI----PSSEASDRIKTGF 124
I L+ + + Q+ AQL TT + G+ P A ++K G+
Sbjct: 298 NTLIRLLK------------TTQIINQVNAQLVTTKNGGLTILAPDDGAFSQLKAGY 432
>CB829244
Length = 478
Score = 31.6 bits (70), Expect = 0.19
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +2
Query: 7 NGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFP 46
+ F S+ +P+ SL V+ +S+PSSSSS+SFP
Sbjct: 104 SSFSFSTTTPSSISLGNTNSSKRCVSFSSTPSSSSSASFP 223
>BP071119
Length = 412
Score = 30.8 bits (68), Expect = 0.32
Identities = 15/35 (42%), Positives = 24/35 (67%)
Frame = -1
Query: 10 YLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSS 44
Y+ +I + +KR +L P V+S+SSP SSS+S+
Sbjct: 133 YMFAILSQQAKVKRTSLIPQPVSSSSSPDSSSNSA 29
>TC14416 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partial (28%)
Length = 695
Score = 28.9 bits (63), Expect = 1.2
Identities = 18/59 (30%), Positives = 29/59 (48%)
Frame = -1
Query: 33 SASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKNYEEAIEELQKLLREKNELKAT 91
S+ SPSS S SS PS+ P+ + PA + T ++ + L ++E+ AT
Sbjct: 485 SS*SPSSPSPSSAPSISASSPLGSQPASVSLTPSAPSSWSSRVSVLSSSCWAQSEVWAT 309
>AV413145
Length = 397
Score = 28.9 bits (63), Expect = 1.2
Identities = 29/119 (24%), Positives = 44/119 (36%), Gaps = 4/119 (3%)
Frame = +1
Query: 11 LSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKN 70
L+ +SPA L+ P ASA P S PS A I+ L K+
Sbjct: 79 LAQLSPASAPLQPAPPVPTTPASAPKPLVPSLPESPS--DSTAPDTAAVDIVGILRKAKS 252
Query: 71 YEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGI----PSSEASDRIKTGFL 125
+ I ++ + + Q+ +QL T + G+ P A +K GFL
Sbjct: 253 FNVLIRLMK------------TTQLINQLNSQLLTIKTGGLTILAPDDSAFSELKAGFL 393
>BP043430
Length = 493
Score = 28.9 bits (63), Expect = 1.2
Identities = 22/58 (37%), Positives = 33/58 (55%)
Frame = +3
Query: 11 LSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMG 68
LSS S +SL + L+ +S+SS SSS+S SFPS P+ + +P+ T +G
Sbjct: 237 LSSSSSLNSSLSLPSSSSLL-SSSSSSSSSTSLSFPSSSLSLPLPSLSSPLALTGILG 407
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 28.9 bits (63), Expect = 1.2
Identities = 24/63 (38%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Frame = -3
Query: 2 STSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFP--SLIQDRPVFAAPA 59
S+S+ + F SS S S + + V S+SS SSSSSSS P S F P
Sbjct: 466 SSSSSSPFSSSSSSSCACSTRDLPRVGCGVCSSSSSSSSSSSSTPWFSFSTSLSFFFLPP 287
Query: 60 PII 62
P+I
Sbjct: 286 PLI 278
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,561,756
Number of Sequences: 28460
Number of extensions: 82685
Number of successful extensions: 1183
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 819
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 993
length of query: 324
length of database: 4,897,600
effective HSP length: 90
effective length of query: 234
effective length of database: 2,336,200
effective search space: 546670800
effective search space used: 546670800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0201.15


