

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.13
(413 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
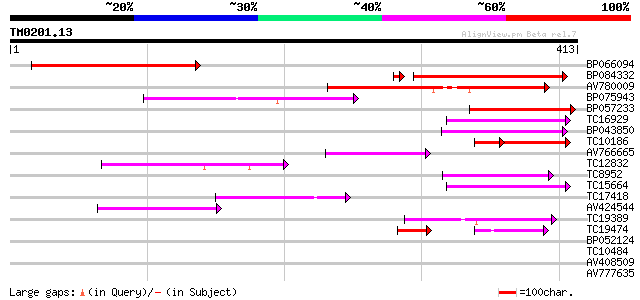
Score E
Sequences producing significant alignments: (bits) Value
BP066094 237 2e-63
BP084332 225 1e-59
AV780009 135 2e-32
BP075943 76 1e-14
BP057233 72 2e-13
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 71 3e-13
BP043850 71 4e-13
TC10186 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprot... 50 2e-11
AV766665 62 1e-10
TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 59 1e-09
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 56 1e-08
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 56 1e-08
TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partia... 53 8e-08
AV424544 52 2e-07
TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 52 2e-07
TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retro... 37 5e-06
BP052124 35 0.030
TC10484 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragm... 28 0.098
AV408509 33 0.11
AV777635 32 0.15
>BP066094
Length = 532
Score = 237 bits (605), Expect = 2e-63
Identities = 116/123 (94%), Positives = 117/123 (94%)
Frame = +2
Query: 17 SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFV 76
SAFLNGYISEEVYVHQPPG EDEK DH+FKLKKSLYGLKQAPRAWYERLSSFLLENE V
Sbjct: 164 SAFLNGYISEEVYVHQPPGXEDEKNSDHIFKLKKSLYGLKQAPRAWYERLSSFLLENEXV 343
Query: 77 RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFL 136
RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN SLCKEFSEMMQAEFEM MMGELKYFL
Sbjct: 344 RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANPSLCKEFSEMMQAEFEMRMMGELKYFL 523
Query: 137 GIQ 139
GIQ
Sbjct: 524 GIQ 532
>BP084332
Length = 368
Score = 225 bits (574), Expect(2) = 1e-59
Identities = 111/112 (99%), Positives = 111/112 (99%)
Frame = +2
Query: 295 QSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQILESNIPIYCDNTAAISLSKNPILHS 354
QSTIALSTAEAEYISAAICSTQMLWMKH LEDYQILESNIPIYCDNTAAISLSKNPILHS
Sbjct: 32 QSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS 211
Query: 355 RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL 406
RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL
Sbjct: 212 RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL 367
Score = 21.6 bits (44), Expect(2) = 1e-59
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +1
Query: 280 CQFLGSNL 287
CQFLGSNL
Sbjct: 4 CQFLGSNL 27
>AV780009
Length = 529
Score = 135 bits (339), Expect = 2e-32
Identities = 69/168 (41%), Positives = 106/168 (63%), Gaps = 6/168 (3%)
Frame = -1
Query: 232 AVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWA 291
A R+LRY+KG GL + S KL Y D+D+AG R+S +G FLG++L+SW
Sbjct: 526 AATRVLRYVKGAPAQGLFFSADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSLISWR 347
Query: 292 SKRQSTIALSTAEAEY--ISAAICSTQMLWMKH*LEDYQILESN----IPIYCDNTAAIS 345
+K+Q+T++ S++EAEY ++A +C Q W+ + +Q L+ N +P++CDN +A+
Sbjct: 346 TKKQTTVSRSSSEAEYRALAATVCEVQ--WLSY---LFQFLKLNVPLPVPLFCDNQSALH 182
Query: 346 LSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKP 393
++ NP H R KHIE+ H +R +Q G++ L + T HQ ADIFTKP
Sbjct: 181 IAHNPTFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTKP 38
>BP075943
Length = 547
Score = 75.9 bits (185), Expect = 1e-14
Identities = 45/159 (28%), Positives = 78/159 (48%), Gaps = 2/159 (1%)
Frame = -3
Query: 98 IYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKE 157
+YVDD++ + + + +F + +GE KYFLG+++ ++ G ++Q KY +
Sbjct: 515 LYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIARSTSGIVLNQRKYALQ 336
Query: 158 LLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKL--YRGMIGSLLYLTASRPDILFSV 215
L+ + H T +G + YR ++G LLYL +RPDI F+V
Sbjct: 335 LISDSGHF-GFQPRFYSHGTNSQTLGTNTGTPLTDIGSYRRIVGRLLYLNTTRPDITFAV 159
Query: 216 HLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTS 254
+ ++F S P + H + L+YL G+ GL Y +S
Sbjct: 158 NQLSQFLSAPTDIHEQQLTGFLKYL*GSPGSGLFYPASS 42
>BP057233
Length = 473
Score = 71.6 bits (174), Expect = 2e-13
Identities = 32/77 (41%), Positives = 51/77 (65%)
Frame = -2
Query: 336 IYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLA 395
++CDN +A +L+ NP+LH+R+KHIE+ H+IRD V + +++ +V T Q AD TKPL+
Sbjct: 445 LWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKPLS 266
Query: 396 EDRFNFILKNLNMDFCP 412
RF+ + L + P
Sbjct: 265 HTRFSQLRDKLGVIHSP 215
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 71.2 bits (173), Expect = 3e-13
Identities = 32/91 (35%), Positives = 54/91 (59%), Gaps = 1/91 (1%)
Frame = +1
Query: 319 WMKH*LEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLL 377
W+ + L+D ++ E +YCDN +A ++ NP+ H R KHIE+ H +R+ +QKG++ L
Sbjct: 4 WLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIHL 183
Query: 378 KFVDTDHQWADIFTKPLAEDRFNFILKNLNM 408
+ + ADI+TK L+ F+ I L +
Sbjct: 184 LPISSSEPLADIYTKALSPQNFHQICAKLGL 276
>BP043850
Length = 515
Score = 70.9 bits (172), Expect = 4e-13
Identities = 34/93 (36%), Positives = 55/93 (58%), Gaps = 1/93 (1%)
Frame = -1
Query: 315 TQMLWMKH*LEDYQILESN-IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKG 373
++++W++ L + L+S P++ DNT+AI ++ NP+ H +HIEV H +R+ +
Sbjct: 515 SEIIWLRGLLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDCHSVREAYDRR 336
Query: 374 VLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL 406
V+ L V T Q ADI TK L R NF++ L
Sbjct: 335 VITLPHVSTSVQIADILTKSLTRQRHNFLVSKL 237
>TC10186 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (4%)
Length = 528
Score = 49.7 bits (117), Expect(2) = 2e-11
Identities = 26/51 (50%), Positives = 32/51 (61%)
Frame = +3
Query: 358 HIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 408
+IE KYHF+RD V KG + LK TD Q ADI TK L +RF + LN+
Sbjct: 69 NIETKYHFLRDQVTKGKISLKHCGTDLQVADIMTKGLKTERFRNMRAMLNV 221
Score = 35.4 bits (80), Expect(2) = 2e-11
Identities = 14/22 (63%), Positives = 18/22 (81%)
Frame = +2
Query: 339 DNTAAISLSKNPILHSRAKHIE 360
DN +AI L+KNP+ H R+KHIE
Sbjct: 5 DNKSAIDLAKNPVSHGRSKHIE 70
>AV766665
Length = 601
Score = 62.4 bits (150), Expect = 1e-10
Identities = 32/76 (42%), Positives = 44/76 (57%)
Frame = +3
Query: 231 TAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSW 290
T RI YLK + G +++K + + G+ ADY G +R ST G FL NLV+W
Sbjct: 360 TFTTRIF*YLKANSRRGPLFQKEGKSSMDGFTYADYLGSIVDRLSTMGYYMFLSGNLVTW 539
Query: 291 ASKRQSTIALSTAEAE 306
SK+Q+ IA S+ EAE
Sbjct: 540 RSKQQNIIARSSGEAE 587
>TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (9%)
Length = 747
Score = 59.3 bits (142), Expect = 1e-09
Identities = 37/145 (25%), Positives = 71/145 (48%), Gaps = 9/145 (6%)
Frame = +2
Query: 68 SFLLENEFVRGKVDTTLFCKTYKD-DILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEM 126
SF++ + R D + K + D D +I+ +YVDD++ N+ +E + EF+M
Sbjct: 8 SFIMSLGYNRLSSDHCTYHKRFDDNDFIILLLYVDDMLVVGPNKDRVQELKAQLAREFDM 187
Query: 127 SMMGELKYFLGIQV--DQTPEGTYIHQSKYTKELLKKFNMLESTVAKTP------MHPTC 178
+G LG+Q+ D+ ++ Q Y +++L++FNM + TP + +
Sbjct: 188 KDLGPANKILGMQIHRDRKDRRIWLSQKNYLQKVLRRFNMQDCNPISTPLPVNYKLSSSM 367
Query: 179 ILEKEDKSGKVCQKLYRGMIGSLLY 203
I E + ++ + Y +GSL+Y
Sbjct: 368 IPSSEAERMEMSRVPYASAVGSLMY 442
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 56.2 bits (134), Expect = 1e-08
Identities = 29/82 (35%), Positives = 48/82 (58%), Gaps = 1/82 (1%)
Frame = +3
Query: 316 QMLWMKH*LEDYQILESN-IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGV 374
+ LW+ + L D +I + IYCDN +A+ L+ N + H R ++IE+ H + V G+
Sbjct: 21 EALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFGI 200
Query: 375 LLLKFVDTDHQWADIFTKPLAE 396
L L V + Q AD+FTK +++
Sbjct: 201 LHLLHVPSSDQVADVFTKTISQ 266
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 56.2 bits (134), Expect = 1e-08
Identities = 27/91 (29%), Positives = 51/91 (55%), Gaps = 1/91 (1%)
Frame = +1
Query: 319 WMKH*LEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLL 377
W+ + L+D ++ S +YCD+ +A ++ N + H R KH+++ H +R+ +Q + L
Sbjct: 13 WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAKLFHL 192
Query: 378 KFVDTDHQWADIFTKPLAEDRFNFILKNLNM 408
+ + Q ADI TKPL F+ ++ L +
Sbjct: 193 LPISSVDQTADILTKPLESGPFSHLVSKLGV 285
>TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partial (20%)
Length = 739
Score = 53.1 bits (126), Expect = 8e-08
Identities = 36/99 (36%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Frame = -1
Query: 151 QSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGK-VCQKLYRGMIGSLLYLTASRP 209
Q Y ++LKKF M S T + I GK V Y+ +IGS+ YL R
Sbjct: 739 QKIYVDDILKKFKMTNSKYISTTIGGKEIEAGRRNGGKRVDSTYYKSLIGSVRYLNTVRS 560
Query: 210 DILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGL 248
DI+ V L +RF +P + H +R LRY+KGT G+
Sbjct: 559 DIVCGVGLRSRFM-EP*DCH*QGAQRSLRYIKGTLKDGI 446
>AV424544
Length = 276
Score = 52.0 bits (123), Expect = 2e-07
Identities = 26/90 (28%), Positives = 49/90 (53%)
Frame = +3
Query: 65 RLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEF 124
+LSS+L +++ D +LF K ++ +YVDD+I + + + + +F
Sbjct: 6 KLSSYLHILGYIQSAHDHSLFTKFRDASFTVILVYVDDLILAGNDLNEIQCVKNKLDIQF 185
Query: 125 EMSMMGELKYFLGIQVDQTPEGTYIHQSKY 154
+ +G LKYFLG++V ++ G ++ Q KY
Sbjct: 186 RIKDLGTLKYFLGLEVARSSCGLFLSQRKY 275
>TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (6%)
Length = 498
Score = 51.6 bits (122), Expect = 2e-07
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 3/114 (2%)
Frame = +3
Query: 288 VSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQILESNIPIYC---DNTAAI 344
V+W S+ Q +ALSTAEAE+I+A ++LWMK+ L++ + PI C A
Sbjct: 27 VAWPSRLQKCVALSTAEAEFIAATEACHELLWMKNFLQNAWF--HSHPILCCIVITKALF 200
Query: 345 SLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDR 398
+L++ + + +RD + +L L+ + TD AD+ TK L ++
Sbjct: 201 TLARILLFIQDPSTLMFVIIGLRDVLNSKLLELEKIHTDDDGADMMTKSLPREK 362
>TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retroelement pol
polyprotein, partial (3%)
Length = 517
Score = 37.0 bits (84), Expect(2) = 5e-06
Identities = 21/54 (38%), Positives = 32/54 (58%)
Frame = -2
Query: 339 DNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTK 392
DN AA+ + K RAKHIE+ +HF+ D + +FV+++ Q D+FTK
Sbjct: 354 DNKAALHMPKI*F-SMRAKHIEIDFHFL*DRRLYLDISTRFVNSNDQLTDVFTK 196
Score = 29.6 bits (65), Expect(2) = 5e-06
Identities = 13/25 (52%), Positives = 16/25 (64%)
Frame = -1
Query: 283 LGSNLVSWASKRQSTIALSTAEAEY 307
L NL+SW SK +A S AEAE+
Sbjct: 517 LSDNLISWKSKETIIVARSRAEAEF 443
>BP052124
Length = 467
Score = 34.7 bits (78), Expect = 0.030
Identities = 14/39 (35%), Positives = 25/39 (63%)
Frame = -2
Query: 337 YCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 375
+CDN +A++L+ PI HSR H EV + ++ + G++
Sbjct: 454 FCDNNSALTLAPRPI*HSRPVHFEVDCPYPKEKLGTGLI 338
>TC10484 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (20%)
Length = 479
Score = 28.5 bits (62), Expect(2) = 0.098
Identities = 10/19 (52%), Positives = 15/19 (78%)
Frame = +3
Query: 332 SNIPIYCDNTAAISLSKNP 350
S +P++CDN AA+ +S NP
Sbjct: 45 SPMPLWCDNQAALHISSNP 101
Score = 23.1 bits (48), Expect(2) = 0.098
Identities = 9/13 (69%), Positives = 9/13 (69%)
Frame = +2
Query: 352 LHSRAKHIEVKYH 364
LH R KHIEV H
Sbjct: 110 LHERTKHIEVDCH 148
>AV408509
Length = 428
Score = 32.7 bits (73), Expect = 0.11
Identities = 31/110 (28%), Positives = 48/110 (43%), Gaps = 5/110 (4%)
Frame = -3
Query: 100 VDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQV-----DQTPEGTYIHQSKY 154
V D I+ N L +E + +FE +G +KYF G QV + E + +++
Sbjct: 324 VIDFIY-MTNDELLEE*KKTTMCQFERLDLGLMKYFDGHQVMAYK*IKNMEKCFFYKASM 148
Query: 155 TKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYL 204
+ + F + V K L +ED KV LYR + SL+YL
Sbjct: 147 LQIYYRSFALCRMGVNKN-------LLREDGKEKVDDTLYRKLGQSLIYL 19
>AV777635
Length = 382
Score = 32.3 bits (72), Expect = 0.15
Identities = 15/35 (42%), Positives = 23/35 (64%)
Frame = -2
Query: 219 ARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT 253
++F DP E HL RIL+YLK + GL+++K+
Sbjct: 381 SQFMHDPHERHLD---RILQYLKASPGRGLLFRKS 286
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,106,849
Number of Sequences: 28460
Number of extensions: 92786
Number of successful extensions: 497
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 492
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 493
length of query: 413
length of database: 4,897,600
effective HSP length: 93
effective length of query: 320
effective length of database: 2,250,820
effective search space: 720262400
effective search space used: 720262400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0201.13


