

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.1
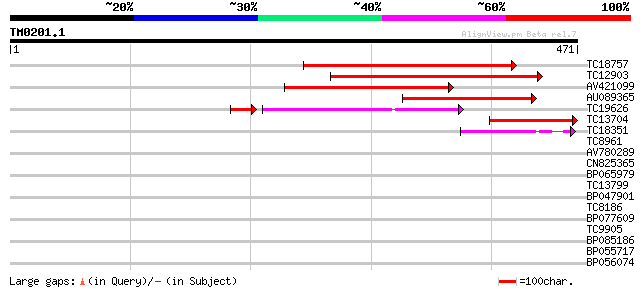
(471 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18757 weakly similar to UP|ALAA_ARATH (Q9LI83) Potential phosp... 253 4e-68
TC12903 weakly similar to UP|ALA9_ARATH (Q9SX33) Potential phosp... 182 8e-47
AV421099 174 2e-44
AU089365 160 5e-40
TC19626 similar to UP|Q7XEQ1 (Q7XEQ1) Contains similarity to chr... 135 3e-38
TC13704 weakly similar to UP|ALA8_ARATH (Q9LK90) Potential phosp... 146 6e-36
TC18351 similar to GB|AAF79467.1|8778459|AC022492 F1L3.21 {Arabi... 75 3e-14
TC8961 similar to GB|BAB71852.1|16902294|AB062739 kinesin-relate... 33 0.077
AV780289 33 0.10
CN825365 30 1.1
BP065979 28 2.5
TC13799 similar to UP|Q8LEY9 (Q8LEY9) UVB-resistance protein-lik... 28 3.2
BP047901 28 3.2
TC8186 similar to UP|ALA3_ARATH (Q9XIE6) Potential phospholipid-... 28 4.2
BP077609 28 4.2
TC9905 27 7.2
BP085186 27 7.2
BP055717 27 9.4
BP056074 27 9.4
>TC18757 weakly similar to UP|ALAA_ARATH (Q9LI83) Potential
phospholipid-transporting ATPase 10 (Aminophospholipid
flippase 10) , partial (15%)
Length = 540
Score = 253 bits (647), Expect = 4e-68
Identities = 118/177 (66%), Positives = 137/177 (76%)
Frame = +2
Query: 245 YMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVIS 304
+MS YNVFFTSLPVIALGVFDQDVS+KLC K+P LY EGV++ LFSW RI GW +NGV S
Sbjct: 5 FMSLYNVFFTSLPVIALGVFDQDVSSKLCFKFPLLYQEGVQNILFSWKRIFGWAINGVSS 184
Query: 305 SLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFI 364
S IFF + +QAFR G+VV EILG T+Y+ VVW VNCQMALSI+YFT+IQHF I
Sbjct: 185 SAIIFFFCIRAFKHQAFREGGEVVGLEILGATLYTCVVWVVNCQMALSISYFTYIQHFVI 364
Query: 365 WGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTY 421
WGSI WYVFL+ YG + P +S+TAY VF EA APS YW+ T+LVVV LLPYF Y
Sbjct: 365 WGSILLWYVFLMAYGAIDPTLSTTAYKVFTEALAPSPSYWVVTLLVVVAALLPYFAY 535
>TC12903 weakly similar to UP|ALA9_ARATH (Q9SX33) Potential
phospholipid-transporting ATPase 9 (Aminophospholipid
flippase 9) , partial (14%)
Length = 746
Score = 182 bits (463), Expect = 8e-47
Identities = 87/176 (49%), Positives = 116/176 (65%)
Frame = +3
Query: 267 DVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQ 326
DVSA+ + F EGV++ LFSW RI+ WMLNG IS++ IFF T ++ QAF +G+
Sbjct: 3 DVSARYASSFLFYIKEGVQNILFSWRRILSWMLNGFISAIIIFFFCTKAMGIQAFDEEGR 182
Query: 327 VVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLSPAIS 386
++L TMY+ VVW VN QMAL+I YFT IQH IWGSI FW++FLL YG L P+IS
Sbjct: 183 TAGKDMLAATMYTCVVWVVNLQMALAIRYFTLIQHVCIWGSIAFWHLFLLAYGALPPSIS 362
Query: 387 STAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVE 442
+ AY VF+E APS + + T V + L+PYF+ Q F PMYH++++ R E
Sbjct: 363 TNAYKVFIETLAPSPSFGMVTFFVAISTLIPYFSCSTIQMWFFPMYHEMVRWTRYE 530
>AV421099
Length = 428
Score = 174 bits (442), Expect = 2e-44
Identities = 76/140 (54%), Positives = 104/140 (74%)
Frame = +3
Query: 229 FEAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTL 288
FEA+A FSGQ Y+DWYM +NV TSLPVI+LGVF+QDV +++CL++P LY +G ++
Sbjct: 3 FEAFAGFSGQSVYDDWYMILFNVVLTSLPVISLGVFEQDVPSEVCLQFPALYQQGPKNLF 182
Query: 289 FSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQ 348
F W RI+GWM NG+ SSL IFFL +Q+FR +GQ D +G TM++ ++W VN Q
Sbjct: 183 FDWYRILGWMGNGLYSSLVIFFLVIIIFYDQSFRSNGQTADMAAVGTTMFTCIIWAVNSQ 362
Query: 349 MALSINYFTWIQHFFIWGSI 368
+AL++ +FTWIQH F+WGSI
Sbjct: 363 IALTMCHFTWIQHLFVWGSI 422
>AU089365
Length = 583
Score = 160 bits (404), Expect = 5e-40
Identities = 69/111 (62%), Positives = 84/111 (75%)
Frame = +2
Query: 327 VVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLSPAIS 386
VV E+LG TMY+ VVW VNCQMALSI+YFT+IQH IWG + FWY+F L YG + P +S
Sbjct: 218 VVGLEVLGATMYTCVVWVVNCQMALSISYFTYIQHLLIWGGVLFWYIFFLXYGAMDPTLS 397
Query: 387 STAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQ 437
+TAY VFVEACAP+ YWL T+LV+V L PYF Y + Q RF PM+H +IQ
Sbjct: 398 TTAYKVFVEACAPAPSYWLITLLVLVASLFPYFVYASIQMRFFPMFHQMIQ 550
>TC19626 similar to UP|Q7XEQ1 (Q7XEQ1) Contains similarity to chromaffin
granule ATPase II homolog, partial (15%)
Length = 576
Score = 135 bits (341), Expect(2) = 3e-38
Identities = 65/167 (38%), Positives = 97/167 (57%)
Frame = +3
Query: 211 MICYFFYKNIAFGFTLFWFEAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSA 270
++ YFFYKN+ F T FWF FSGQ Y+DW+ S YNV FT+LPVI +G+FD+DVSA
Sbjct: 78 VVIYFFYKNLTFTLTQFWFTFQTGFSGQRFYDDWFQSLYNVIFTALPVIIVGLFDKDVSA 257
Query: 271 KLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDF 330
L KYP LY+EG+++ F W + W V SL ++ + S ++ A G++
Sbjct: 258 SLSKKYPELYMEGIKNVFFKWKVVAIWAFFSVYQSLIFYYYVSTSNLS-AKNSAGKIFGL 434
Query: 331 EILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFLLV 377
+ ++ VV TVN ++ L N T + + GSI W++F+ +
Sbjct: 435 WDVSTMAFTCVVVTVNLRLLLICNSITRWHYISVGGSILAWFLFIFI 575
Score = 39.7 bits (91), Expect(2) = 3e-38
Identities = 17/22 (77%), Positives = 18/22 (81%)
Frame = +1
Query: 184 DFAIAQFRFLERLLLVHGHWCY 205
DFAIAQFR+L LLLVHG W Y
Sbjct: 1 DFAIAQFRYLADLLLVHGRWSY 66
>TC13704 weakly similar to UP|ALA8_ARATH (Q9LK90) Potential
phospholipid-transporting ATPase 8 (Aminophospholipid
flippase 8) , partial (3%)
Length = 517
Score = 146 bits (369), Expect = 6e-36
Identities = 71/73 (97%), Positives = 72/73 (98%)
Frame = +2
Query: 399 PSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQG 458
PSAVYWLATVLVVVCVLLPYFTYRAF SRFLPMYHDIIQRKRVEGFEVEISDELPTQVQG
Sbjct: 2 PSAVYWLATVLVVVCVLLPYFTYRAFXSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQG 181
Query: 459 KLMHMRERLKQRE 471
KLMHMRERLK+RE
Sbjct: 182 KLMHMRERLKRRE 220
>TC18351 similar to GB|AAF79467.1|8778459|AC022492 F1L3.21 {Arabidopsis
thaliana;} , partial (8%)
Length = 619
Score = 74.7 bits (182), Expect = 3e-14
Identities = 40/96 (41%), Positives = 56/96 (57%)
Frame = +1
Query: 375 LLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHD 434
L++YG LSP S TAY +FVE AP+ +YWL T+LV +LPY + +FQ F PM H
Sbjct: 1 LILYGMLSPTYSKTAYQIFVEVLAPAPIYWLTTLLVTFTCILPYLAHISFQRCFNPMDHH 180
Query: 435 IIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQR 470
IIQ ++ ++ ++ D+ M RER K R
Sbjct: 181 IIQ--EIKYYKKDVEDQ--------RMWTRERSKAR 258
>TC8961 similar to GB|BAB71852.1|16902294|AB062739 kinesin-related protein
{Arabidopsis thaliana;} , partial (8%)
Length = 735
Score = 33.5 bits (75), Expect = 0.077
Identities = 14/30 (46%), Positives = 18/30 (59%)
Frame = +3
Query: 193 LERLLLVHGHWCYRRISLMICYFFYKNIAF 222
L LL HGHWC ISL++C F + I +
Sbjct: 429 LIELLYPHGHWCNSFISLLVCKLFKRYIDY 518
>AV780289
Length = 445
Score = 33.1 bits (74), Expect = 0.10
Identities = 14/39 (35%), Positives = 24/39 (60%)
Frame = -1
Query: 145 TGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVMAS 183
T I++IGDG ND+ ML+ A +G+ ++ ++AS
Sbjct: 415 TAHEIMAIGDGENDIEMLELASLGIALTNGSERTKLVAS 299
>CN825365
Length = 511
Score = 29.6 bits (65), Expect = 1.1
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Frame = +1
Query: 412 VCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEGF---EVEISDELPTQVQGKLMHMRER 466
+CV L F Y+ F + FL ++ I K++E + + I+ EL G + H+R +
Sbjct: 52 ICVQLFLFVYKLFSANFLLVFPPFISGKKIETWGSKGLAITVELQAHHFGVMGHLRSQ 225
>BP065979
Length = 362
Score = 28.5 bits (62), Expect = 2.5
Identities = 26/82 (31%), Positives = 41/82 (49%), Gaps = 3/82 (3%)
Frame = -1
Query: 393 FVEACAPSAVYWLA---TVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEGFEVEIS 449
+VEA P+ +YW A L + ++LP TY + + PM H IIQ ++ ++ +I
Sbjct: 344 WVEALGPAPIYWSAHFWLQLHAIFLILP--TYHS-KDVSTPMDHHIIQ--EIKYYKKDIE 180
Query: 450 DELPTQVQGKLMHMRERLKQRE 471
D+ M RER K R+
Sbjct: 179 DQ--------HMWTRERSKARQ 138
>TC13799 similar to UP|Q8LEY9 (Q8LEY9) UVB-resistance protein-like, partial
(16%)
Length = 458
Score = 28.1 bits (61), Expect = 3.2
Identities = 11/30 (36%), Positives = 18/30 (59%)
Frame = -3
Query: 370 FWYVFLLVYGYLSPAISSTAYMVFVEACAP 399
F+YVF VYGY+ P++ + M+ + P
Sbjct: 417 FFYVFGXVYGYI*PSMPNAITMLIINVFVP 328
>BP047901
Length = 415
Score = 28.1 bits (61), Expect = 3.2
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 4/56 (7%)
Frame = -3
Query: 365 WGSIFFWYVFLLVYGYLSPAISSTAYMVFVE---ACAPSAVYWL-ATVLVVVCVLL 416
WG +FF V +LVY S + + ++ +E +CA ++WL ++L+ C+ L
Sbjct: 341 WGLVFFTLVLVLVYECAS--LDNLVQLLLLEDHVSCAVMGMHWLDFSLLLEGCIYL 180
>TC8186 similar to UP|ALA3_ARATH (Q9XIE6) Potential
phospholipid-transporting ATPase 3 (Aminophospholipid
flippase 3) , partial (10%)
Length = 804
Score = 27.7 bits (60), Expect = 4.2
Identities = 18/66 (27%), Positives = 29/66 (43%), Gaps = 1/66 (1%)
Frame = +1
Query: 373 VFLLVYGYLSPAISSTAYMVFVEACAPSAVY-WLATVLVVVCVLLPYFTYRAFQSRFLPM 431
+F+L+ +S + FV + +Y +L +LV L F Y+ Q F P
Sbjct: 1 IFVLIDSVISTPYDRQENVYFVIYVLMNTLYFYLTLILVPAAALFGDFVYQGVQRWFFPY 180
Query: 432 YHDIIQ 437
+ IIQ
Sbjct: 181 DYQIIQ 198
>BP077609
Length = 311
Score = 27.7 bits (60), Expect = 4.2
Identities = 19/58 (32%), Positives = 28/58 (47%)
Frame = +1
Query: 73 GISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSCASVICCRSS 130
GIS+ K SS + T + II+G ++N+ F LA SC ++CC S
Sbjct: 157 GISRATQCK*SSR--QPTKIWPAIING-------DENISPCKFSLANSCTPLVCCSLS 303
>TC9905
Length = 549
Score = 26.9 bits (58), Expect = 7.2
Identities = 9/18 (50%), Positives = 11/18 (61%)
Frame = -2
Query: 203 WCYRRISLMICYFFYKNI 220
WC +SL CYF YK +
Sbjct: 332 WCKNFVSLPPCYFLYKEV 279
>BP085186
Length = 388
Score = 26.9 bits (58), Expect = 7.2
Identities = 15/56 (26%), Positives = 24/56 (42%)
Frame = +2
Query: 327 VVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLS 382
V F LGV ++ L I +F W FF + FF++ F + ++S
Sbjct: 134 VFSFSFLGVDIF------------LFILFFLWFCFFFFFFFFFFFFFFFFFFFFIS 265
>BP055717
Length = 339
Score = 26.6 bits (57), Expect = 9.4
Identities = 17/62 (27%), Positives = 28/62 (44%), Gaps = 3/62 (4%)
Frame = -2
Query: 327 VVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQHFFIWGSIFFWYVFLLV---YGYLSP 383
++DFE+ ++YSI +FTW + F F W+ +L + + Y SP
Sbjct: 221 ILDFEVFSFSLYSI------------NQFFTWFXYNFRRLF*FHWF*YLWLPTRFPYASP 78
Query: 384 AI 385
I
Sbjct: 77 CI 72
>BP056074
Length = 485
Score = 26.6 bits (57), Expect = 9.4
Identities = 9/18 (50%), Positives = 11/18 (61%)
Frame = -3
Query: 416 LPYFTYRAFQSRFLPMYH 433
LPYF Y + R P+YH
Sbjct: 483 LPYFAYSSIPMRVFPLYH 430
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,482,282
Number of Sequences: 28460
Number of extensions: 128771
Number of successful extensions: 1136
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1124
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1133
length of query: 471
length of database: 4,897,600
effective HSP length: 94
effective length of query: 377
effective length of database: 2,222,360
effective search space: 837829720
effective search space used: 837829720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0201.1


