

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0200.8
(213 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
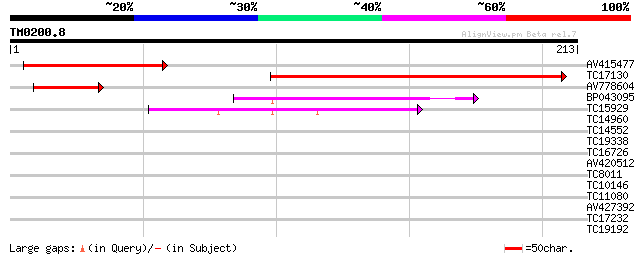
Score E
Sequences producing significant alignments: (bits) Value
AV415477 104 1e-23
TC17130 similar to UP|AAR24679 (AAR24679) At3g50960, partial (36%) 92 7e-20
AV778604 48 1e-06
BP043095 43 5e-05
TC15929 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, ... 41 1e-04
TC14960 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%) 38 0.002
TC14552 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, ... 36 0.004
TC19338 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m ... 35 0.010
TC16726 similar to UP|GS11_ARATH (Q9LMP7) Golgi SNARE 11 protein... 27 2.0
AV420512 27 2.7
TC8011 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomera... 27 3.5
TC10146 similar to PIR|T06666|T06666 26S proteasome regulatory p... 26 4.5
TC11080 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin r... 26 4.5
AV427392 25 7.8
TC17232 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific ... 25 7.8
TC19192 weakly similar to UP|PSD6_ORYSA (Q8W425) 26S proteasome ... 25 7.8
>AV415477
Length = 165
Score = 104 bits (260), Expect = 1e-23
Identities = 54/54 (100%), Positives = 54/54 (100%)
Frame = +3
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRN 59
IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRN
Sbjct: 3 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRN 164
>TC17130 similar to UP|AAR24679 (AAR24679) At3g50960, partial (36%)
Length = 549
Score = 92.0 bits (227), Expect = 7e-20
Identities = 44/111 (39%), Positives = 69/111 (61%)
Frame = +1
Query: 99 CKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDE 158
CK+ DKHL L+ +HI+T+F+K++AE +PF KL++ LP + L + D +VGF +
Sbjct: 10 CKIMDKHLKDLSPKHIDTKFIKLDAENAPFFVAKLQVKTLPCIILFRQGVAFDRLVGFQD 189
Query: 159 LGGTDDFSTEDLEERLAKAQVIFLEGESSIHQARSTAQSKRSVRQATRADS 209
LGG DDF+T+ LE L K +I + + +A ++R+VR + ADS
Sbjct: 190 LGGKDDFTTKKLEAMLIKKGIIDDKKNQNDEEAEYDESARRTVRTSVAADS 342
>AV778604
Length = 545
Score = 48.1 bits (113), Expect = 1e-06
Identities = 24/26 (92%), Positives = 25/26 (95%)
Frame = -3
Query: 10 IEKQVLTVVQAVEDKIDDEISALDRL 35
IEKQVLT+VQAVEDKIDDEI ALDRL
Sbjct: 471 IEKQVLTLVQAVEDKIDDEIPALDRL 394
>BP043095
Length = 396
Score = 42.7 bits (99), Expect = 5e-05
Identities = 23/94 (24%), Positives = 48/94 (50%), Gaps = 2/94 (2%)
Frame = +1
Query: 85 SDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLA 142
S +++ F +W PC+ +LG LAK++ F+K++ ++ +A+ + +PT
Sbjct: 139 SKKLIVVDFTASWCGPCRFIAPYLGELAKKYTNVIFLKVDVDELKSVAQDWAVEAMPTFM 318
Query: 143 LIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAK 176
+K + D VVG + E+L++++ K
Sbjct: 319 FVKEGSIVDKVVGAKK---------EELQQKIEK 393
>TC15929 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (49%)
Length = 1406
Score = 41.2 bits (95), Expect = 1e-04
Identities = 31/108 (28%), Positives = 53/108 (48%), Gaps = 5/108 (4%)
Frame = +1
Query: 53 KMAEKRNRWISLGHGDYSELPSEKD--FFSVVKASDRVVCHFFRENW--PCKVADKHLGV 108
K A +R R + E+ S D + S+V SD V F W PC++ +
Sbjct: 247 KFAPRRGRVVCEAQNTAVEVTSITDANWQSLVIESDTAVLVEFWAPWCGPCRMIHPVIDE 426
Query: 109 LAKQHI-ETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVG 155
LAK++ + + K+N ++SP A + I +PT+ + K+ + D V+G
Sbjct: 427 LAKEYAGKLKCYKLNTDESPSTATRYGIRSIPTVIIFKDGEKKDAVIG 570
>TC14960 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%)
Length = 688
Score = 37.7 bits (86), Expect = 0.002
Identities = 21/82 (25%), Positives = 39/82 (46%), Gaps = 2/82 (2%)
Frame = +1
Query: 76 KDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKL 133
K+ S +++ F +W PC+ L +AK+ F+K++ ++ +AE+
Sbjct: 130 KEHLEKGNVSKKLIVVDFTASWCGPCRFIAPILAEIAKKLPHVVFLKVDVDELKTVAEEW 309
Query: 134 KITVLPTLALIKNAKVDDYVVG 155
+ +PT +K K D VVG
Sbjct: 310 NVEAMPTFLFLKEGKAVDKVVG 375
>TC14552 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (55%)
Length = 1016
Score = 36.2 bits (82), Expect = 0.004
Identities = 23/85 (27%), Positives = 46/85 (54%), Gaps = 3/85 (3%)
Frame = +1
Query: 74 SEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLA 130
++ ++ S+V S+ V F W PC++ + LAKQ+ + + K+N ++SP A
Sbjct: 313 TDANWQSLVLESEFPVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDESPSTA 492
Query: 131 EKLKITVLPTLALIKNAKVDDYVVG 155
+ I +PT+ + ++ + D V+G
Sbjct: 493 TRYGIRSIPTVMIFRDGEKKDTVIG 567
>TC19338 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (65%)
Length = 476
Score = 35.0 bits (79), Expect = 0.010
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 3/85 (3%)
Frame = +2
Query: 74 SEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLA 130
+E + +V AS+ V F W PC++ + LAK++ + K+N + SP +A
Sbjct: 44 TESSWNDLVIASEIPVLVDFWAPWCGPCRMIAPLIDELAKEYAGKIACYKLNTDDSPNIA 223
Query: 131 EKLKITVLPTLALIKNAKVDDYVVG 155
+ I +PT+ KN + + V+G
Sbjct: 224 TQYGIRSIPTVLFFKNGEKKESVIG 298
>TC16726 similar to UP|GS11_ARATH (Q9LMP7) Golgi SNARE 11 protein (AtGOS11)
(Golgi SNAP receptor complex member 1-1), partial (80%)
Length = 655
Score = 27.3 bits (59), Expect = 2.0
Identities = 15/59 (25%), Positives = 29/59 (48%)
Frame = +2
Query: 10 IEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHGD 68
+E Q+ + + + +SA +DLES E+ L Q++++ + W+S G D
Sbjct: 167 LEAQLDERMSSFRKLVSASVSAKTDAAENDLESWIEQLLKQLQQVNSQMQAWVSSGGTD 343
>AV420512
Length = 423
Score = 26.9 bits (58), Expect = 2.7
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 5/68 (7%)
Frame = +3
Query: 81 VVKASDRVVCHFFRENWPCKVADK---HLGVLAKQHIETRFVKINAEKSPF--LAEKLKI 135
V D V H+F+ NW +V K H L + + VK++ +EKLK
Sbjct: 36 VTGTEDHRVRHYFKRNWTFEVIRKMRSHYQTLEQNEEASILVKVSVRVYEVFGFSEKLKF 215
Query: 136 TVLPTLAL 143
T LP L +
Sbjct: 216 T-LPNLQI 236
>TC8011 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (89%)
Length = 1305
Score = 26.6 bits (57), Expect = 3.5
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 2/80 (2%)
Frame = +3
Query: 8 EVIEKQVLTVVQAVEDKIDD--EISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
+V+ + V++ I D E+SA + S E + Q+K K W+ LG
Sbjct: 360 DVVVAPPFVYIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVK---WVILG 530
Query: 66 HGDYSELPSEKDFFSVVKAS 85
H + + EKD F KA+
Sbjct: 531 HSERRHIIGEKDEFIGKKAA 590
>TC10146 similar to PIR|T06666|T06666 26S proteasome regulatory particle
chain RPN7 homolog F6I7.30 - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (35%)
Length = 725
Score = 26.2 bits (56), Expect = 4.5
Identities = 12/35 (34%), Positives = 21/35 (59%)
Frame = +3
Query: 16 TVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQ 50
T++ ++ KIDDE+ LD +D E+L E + +
Sbjct: 354 TLLDSMRAKIDDELKKLDEKIADAEENLGESEVRE 458
>TC11080 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin response
modulator, partial (33%)
Length = 556
Score = 26.2 bits (56), Expect = 4.5
Identities = 14/48 (29%), Positives = 24/48 (49%)
Frame = -1
Query: 21 VEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHGD 68
+ + + DE + L+ LD+D L L E H ++ + R RW G +
Sbjct: 499 IGEAVADEAAELE-LDADGLSKLSELPTHLLQGVGVVRLRWSDSGEAE 359
>AV427392
Length = 366
Score = 25.4 bits (54), Expect = 7.8
Identities = 12/49 (24%), Positives = 23/49 (46%)
Frame = +1
Query: 99 CKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNA 147
CK + LA+ + + +F+ +N E+ + L + VLP + A
Sbjct: 55 CKALHPKICQLAEMNPDVQFLHVNYEEHKSMCYSLNVHVLPFFRFYRGA 201
>TC17232 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein-2,
partial (33%)
Length = 966
Score = 25.4 bits (54), Expect = 7.8
Identities = 9/23 (39%), Positives = 14/23 (60%)
Frame = -2
Query: 78 FFSVVKASDRVVCHFFRENWPCK 100
FF + + +VC F R+NW C+
Sbjct: 266 FFQLFQ*EWYLVCDFARQNWDCR 198
>TC19192 weakly similar to UP|PSD6_ORYSA (Q8W425) 26S proteasome non-ATPase
regulatory subunit 6 (26S proteasome regulatory particle
non-ATPase subunit 7) (OsRPN7), partial (34%)
Length = 433
Score = 25.4 bits (54), Expect = 7.8
Identities = 11/35 (31%), Positives = 21/35 (59%)
Frame = +2
Query: 16 TVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQ 50
T++ ++ KIDDE+ +D +D E+L E + +
Sbjct: 224 TLLDSMRAKIDDELKKIDEKIADAEENLGESEVRE 328
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,681,983
Number of Sequences: 28460
Number of extensions: 30483
Number of successful extensions: 158
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 158
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 158
length of query: 213
length of database: 4,897,600
effective HSP length: 86
effective length of query: 127
effective length of database: 2,450,040
effective search space: 311155080
effective search space used: 311155080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0200.8


