

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0195a.7
(69 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
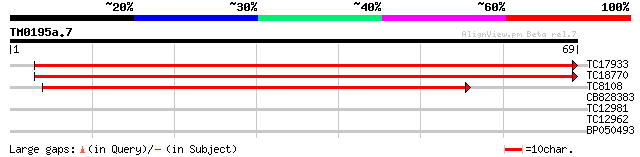
Score E
Sequences producing significant alignments: (bits) Value
TC17933 homologue to UP|CAF02296 (CAF02296) Rho GDP dissociation... 122 1e-29
TC18770 similar to UP|Q7Y0Q8 (Q7Y0Q8) Rac GDP-dissociation inhib... 100 7e-23
TC8108 homologue to UP|CAF02295 (CAF02295) Rho GDP dissociation ... 68 4e-13
CB828383 24 4.8
TC12981 24 4.8
TC12962 similar to AAQ89629 (AAQ89629) At3g50330, partial (27%) 23 8.2
BP050493 23 8.2
>TC17933 homologue to UP|CAF02296 (CAF02296) Rho GDP dissociation inhibitor
2, partial (34%)
Length = 482
Score = 122 bits (306), Expect = 1e-29
Identities = 55/66 (83%), Positives = 61/66 (92%)
Frame = +1
Query: 4 VTVDSSKEMIGTFSPQTDPYTHEMPDETTPSGIFARRAYSARSKFVDDDNKCYLEIDYTF 63
+ VDS+KEMIGTFSPQ +PYTHEMP+ETTPSGIFAR YSAR+KFVDDDNK YLEI+YTF
Sbjct: 43 IKVDSTKEMIGTFSPQAEPYTHEMPEETTPSGIFARGTYSARTKFVDDDNKMYLEINYTF 222
Query: 64 DIRKDW 69
DIRKDW
Sbjct: 223 DIRKDW 240
>TC18770 similar to UP|Q7Y0Q8 (Q7Y0Q8) Rac GDP-dissociation inhibitor 1,
partial (66%)
Length = 948
Score = 100 bits (248), Expect = 7e-23
Identities = 42/66 (63%), Positives = 56/66 (84%)
Frame = +1
Query: 4 VTVDSSKEMIGTFSPQTDPYTHEMPDETTPSGIFARRAYSARSKFVDDDNKCYLEIDYTF 63
V VD++K M+GT+SPQ +PYT+E+ +ETTPSG+FAR YSAR+KFVDDD KCYL+ Y F
Sbjct: 643 VRVDNAKRMLGTYSPQQEPYTYELEEETTPSGLFARGTYSARTKFVDDDRKCYLDATYHF 822
Query: 64 DIRKDW 69
+I+K+W
Sbjct: 823 EIQKNW 840
>TC8108 homologue to UP|CAF02295 (CAF02295) Rho GDP dissociation inhibitor
1, partial (28%)
Length = 599
Score = 67.8 bits (164), Expect = 4e-13
Identities = 28/52 (53%), Positives = 39/52 (74%)
Frame = +2
Query: 5 TVDSSKEMIGTFSPQTDPYTHEMPDETTPSGIFARRAYSARSKFVDDDNKCY 56
+VD SK M+GTF+PQ +PY + + ++TTPSG AR YSA+ KF DDD +C+
Sbjct: 191 SVDQSKGMLGTFAPQKEPYVYALKEDTTPSGALARGVYSAKLKFEDDDQRCH 346
>CB828383
Length = 372
Score = 24.3 bits (51), Expect = 4.8
Identities = 10/22 (45%), Positives = 13/22 (58%)
Frame = -2
Query: 13 IGTFSPQTDPYTHEMPDETTPS 34
+G F P P+T +P ET PS
Sbjct: 356 LGRFDP*FWPHTQSLPVETVPS 291
>TC12981
Length = 614
Score = 24.3 bits (51), Expect = 4.8
Identities = 11/21 (52%), Positives = 12/21 (56%)
Frame = -2
Query: 20 TDPYTHEMPDETTPSGIFARR 40
T+ TH P T PSGI RR
Sbjct: 463 TNSNTHSAPSSTYPSGIRHRR 401
>TC12962 similar to AAQ89629 (AAQ89629) At3g50330, partial (27%)
Length = 545
Score = 23.5 bits (49), Expect = 8.2
Identities = 14/38 (36%), Positives = 18/38 (46%), Gaps = 2/38 (5%)
Frame = +2
Query: 8 SSKEMIGTFSPQT--DPYTHEMPDETTPSGIFARRAYS 43
SS I +P T +TH P TTPS F + Y+
Sbjct: 167 SSATSINNNNPPTPWSTFTHHFPSSTTPS--FPNKLYT 274
>BP050493
Length = 549
Score = 23.5 bits (49), Expect = 8.2
Identities = 7/16 (43%), Positives = 12/16 (74%)
Frame = -3
Query: 54 KCYLEIDYTFDIRKDW 69
KCY E+++ ++RK W
Sbjct: 166 KCY*EMNFKANVRKSW 119
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 971,487
Number of Sequences: 28460
Number of extensions: 8528
Number of successful extensions: 39
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39
length of query: 69
length of database: 4,897,600
effective HSP length: 45
effective length of query: 24
effective length of database: 3,616,900
effective search space: 86805600
effective search space used: 86805600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 48 (23.1 bits)
Lotus: description of TM0195a.7


