

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
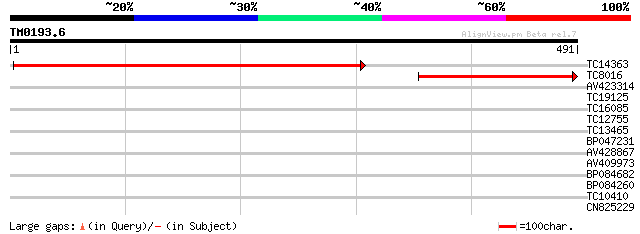
Query= TM0193.6
(491 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14363 homologue to UP|CAT4_SOYBN (O48561) Catalase 4 , partia... 645 0.0
TC8016 homologue to UP|CAT4_SOYBN (O48561) Catalase 4 , partial... 298 1e-81
AV423314 32 0.31
TC19125 UP|GBA1_LOTJA (P49082) Guanine nucleotide-binding protei... 29 1.5
TC16085 28 3.4
TC12755 similar to UP|Q9C587 (Q9C587) Replication factor C large... 27 5.8
TC13465 similar to UP|O23110 (O23110) AP2 domain containing prot... 27 7.6
BP047231 27 7.6
AV428867 27 7.6
AV409973 27 7.6
BP084682 27 9.9
BP084260 27 9.9
TC10410 weakly similar to UP|Q8H5W8 (Q8H5W8) OJ1123_B01.10 prote... 27 9.9
CN825229 27 9.9
>TC14363 homologue to UP|CAT4_SOYBN (O48561) Catalase 4 , partial (63%)
Length = 1045
Score = 645 bits (1664), Expect = 0.0
Identities = 304/305 (99%), Positives = 305/305 (99%)
Frame = +3
Query: 4 QHRPSSAFNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVV 63
+HRPSSAFNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVV
Sbjct: 129 KHRPSSAFNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVV 308
Query: 64 HARGASAKGFFEVTHDVSHLTCADFLRAPGVQTPIILRFSTVIHERGSPETLRDPRGFAV 123
HARGASAKGFFEVTHDVSHLTCADFLRAPGVQTPIILRFSTVIHERGSPETLRDPRGFAV
Sbjct: 309 HARGASAKGFFEVTHDVSHLTCADFLRAPGVQTPIILRFSTVIHERGSPETLRDPRGFAV 488
Query: 124 KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHFPESLH 183
KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHFPESLH
Sbjct: 489 KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHFPESLH 668
Query: 184 MFTFLFDDVGVPQDYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAIKVG 243
MFTFLFDDVGVPQDYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAIKVG
Sbjct: 669 MFTFLFDDVGVPQDYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAIKVG 848
Query: 244 GSNHSHATQDLYESIAAGNYPEWKLFIQTIDPDHEDRFDFDPLDVTKTWPEDIIPLQPVG 303
GSNHSHATQDLYESIAAGNYPEWKLFIQTIDPDHEDRFDFDPLDVTKTWPEDIIPLQPVG
Sbjct: 849 GSNHSHATQDLYESIAAGNYPEWKLFIQTIDPDHEDRFDFDPLDVTKTWPEDIIPLQPVG 1028
Query: 304 RMVLN 308
RMVLN
Sbjct: 1029RMVLN 1043
>TC8016 homologue to UP|CAT4_SOYBN (O48561) Catalase 4 , partial (28%)
Length = 727
Score = 298 bits (763), Expect = 1e-81
Identities = 137/137 (100%), Positives = 137/137 (100%)
Frame = +1
Query: 355 LGPNYLQLPANAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTG 414
LGPNYLQLPANAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTG
Sbjct: 1 LGPNYLQLPANAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTG 180
Query: 415 SRERCAIEKENNFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWISYWSQAD 474
SRERCAIEKENNFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWISYWSQAD
Sbjct: 181 SRERCAIEKENNFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWISYWSQAD 360
Query: 475 RSLGQKIASHLNLRPSI 491
RSLGQKIASHLNLRPSI
Sbjct: 361 RSLGQKIASHLNLRPSI 411
>AV423314
Length = 498
Score = 31.6 bits (70), Expect = 0.31
Identities = 22/71 (30%), Positives = 31/71 (42%)
Frame = +2
Query: 11 FNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVVHARGASA 70
+ S WT +G +W N T G+RG HL+++ A F R VH + A
Sbjct: 230 WTSRMWTKYTGVLIWKTQNPWT-GLRGQFY---DHLLDQTAGFYGCRCAAEPVHVQLNLA 397
Query: 71 KGFFEVTHDVS 81
F EV + S
Sbjct: 398 TYFIEVVNTTS 430
>TC19125 UP|GBA1_LOTJA (P49082) Guanine nucleotide-binding protein alpha-1
subunit (GP-alpha-1), complete
Length = 1597
Score = 29.3 bits (64), Expect = 1.5
Identities = 23/84 (27%), Positives = 33/84 (38%)
Frame = +2
Query: 367 PKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTGSRERCAIEKENN 426
P C H+ FM +HR + NY P++ D V +A + T
Sbjct: 494 PDCTHY------FMENLHRLSDANYVPTK-DDVLYAR-------VRTTGVVEIQFSPVGE 631
Query: 427 FKQPGERYRSFAPDRQDRFVRRWV 450
K+ GE YR F Q R+W+
Sbjct: 632 NKKSGEVYRLFDVGGQRNERRKWI 703
>TC16085
Length = 501
Score = 28.1 bits (61), Expect = 3.4
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = -1
Query: 65 ARGASAKGFFEVTHDVSHLTC 85
ARG S K F V D+ HLTC
Sbjct: 234 ARGTSLKWHFRVLKDMQHLTC 172
>TC12755 similar to UP|Q9C587 (Q9C587) Replication factor C large
subunit-like protein, partial (3%)
Length = 477
Score = 27.3 bits (59), Expect = 5.8
Identities = 10/23 (43%), Positives = 12/23 (51%)
Frame = -2
Query: 390 NYFPSRYDPVRHAERYPIPPAIC 412
N PS Y P RH+ P P +C
Sbjct: 413 NQLPSSYQPPRHSHHLPHPHQLC 345
>TC13465 similar to UP|O23110 (O23110) AP2 domain containing protein RAP2.8
(Fragment), partial (18%)
Length = 609
Score = 26.9 bits (58), Expect = 7.6
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = +1
Query: 356 GPNYLQLPANAPKCAHHNNHH 376
G N L+LP + +HNNHH
Sbjct: 238 GVNILKLPGSDANANNHNNHH 300
>BP047231
Length = 476
Score = 26.9 bits (58), Expect = 7.6
Identities = 17/53 (32%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = -1
Query: 284 DPLDVTKTWPEDIIPLQPVGRMVLNKNIDNFFAENEQLAFC-PAIVVPGISYS 335
DP++V K + + + LQP G ++I + LA C P PG +YS
Sbjct: 230 DPINVMKVFEKSVFCLQPPGDSYTRRSI-----FDSMLAGCVPVFFHPGTAYS 87
>AV428867
Length = 628
Score = 26.9 bits (58), Expect = 7.6
Identities = 15/52 (28%), Positives = 27/52 (51%)
Frame = +1
Query: 417 ERCAIEKENNFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWIS 468
+R + + + F +P +S + + DRF D +S+PR + + RS IS
Sbjct: 217 DRFSEPRGDRFSEPSGSTQSRSAAKGDRFSEPRGDRISEPRDSRQSRSATIS 372
>AV409973
Length = 415
Score = 26.9 bits (58), Expect = 7.6
Identities = 11/34 (32%), Positives = 15/34 (43%)
Frame = +2
Query: 455 DPRVTHEIRSTWISYWSQADRSLGQKIASHLNLR 488
D H +S W+S+W+ SHL LR
Sbjct: 191 DAPAMHGSKSAWMSHWANTSCKSATPARSHLTLR 292
>BP084682
Length = 236
Score = 26.6 bits (57), Expect = 9.9
Identities = 8/18 (44%), Positives = 14/18 (77%)
Frame = -1
Query: 358 NYLQLPANAPKCAHHNNH 375
++L+LP+ A C HH++H
Sbjct: 188 HHLELPSAASSCGHHHSH 135
>BP084260
Length = 333
Score = 26.6 bits (57), Expect = 9.9
Identities = 13/44 (29%), Positives = 22/44 (49%)
Frame = +2
Query: 197 DYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAI 240
+YRH ++Y L + +VY+ W P C + C +E+ I
Sbjct: 152 EYRH------SSYRLSSSPLLLVYIYESWIPHCRIHCRKQEKCI 265
>TC10410 weakly similar to UP|Q8H5W8 (Q8H5W8) OJ1123_B01.10 protein, partial
(12%)
Length = 431
Score = 26.6 bits (57), Expect = 9.9
Identities = 26/112 (23%), Positives = 43/112 (38%), Gaps = 5/112 (4%)
Frame = +1
Query: 357 PNYLQLPANAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPV--RHAERYPIPPAICTG 414
P L P N HH NHH + HR PS++ P+ + + P I T
Sbjct: 34 PPPLGAPLNPHNLNHHYNHHLLSLKLHHRQP----LPSQHHPILPKTLTQPQFQPQIQTQ 201
Query: 415 SRERCAIEKENNFK--QPGERYRSFA-PDRQDRFVRRWVDALSDPRVTHEIR 463
++ + I ++ + P + + A P +RR + R +H R
Sbjct: 202 TQSKSKILTQSTTQTLDPLQLLHNLALPPHSTELLRRSSSSRRISRTSHRFR 357
>CN825229
Length = 641
Score = 26.6 bits (57), Expect = 9.9
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 6/100 (6%)
Frame = +3
Query: 296 IIPLQPVG-RMVLNKNIDNFFAENEQLAFCPA-IVVPGISYSDDKMLQT----RIFSYAD 349
+ L P G R+V++ D + E E L A +++ +SY++DK+ + R+F Y
Sbjct: 186 VYKLLPQGTRLVVSVRGDEPYEELEILKGVDATMLLHDLSYAEDKVSRVHAARRLFEY-- 359
Query: 350 SQRHRLGPNYLQLPANAPKCAHHNNHHEGFMNFMHRDEEV 389
L N L P HH F N +HRD+ V
Sbjct: 360 -----LSDNSLNFPVI---------HHIQFPNGIHRDDLV 437
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,842,934
Number of Sequences: 28460
Number of extensions: 154034
Number of successful extensions: 718
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 704
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 716
length of query: 491
length of database: 4,897,600
effective HSP length: 94
effective length of query: 397
effective length of database: 2,222,360
effective search space: 882276920
effective search space used: 882276920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0193.6


