

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.5
(903 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
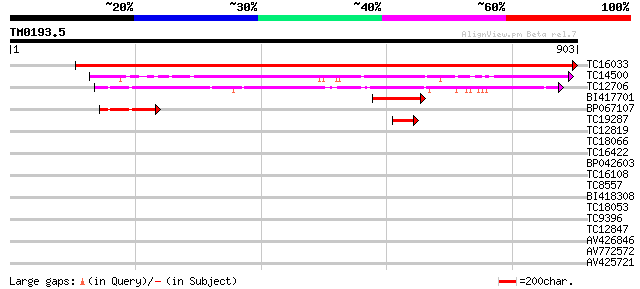
Score E
Sequences producing significant alignments: (bits) Value
TC16033 UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (88%) 1599 0.0
TC14500 UP|Q9S7B1 (Q9S7B1) Nodule INCEPTION protein, complete 521 e-148
TC12706 UP|Q7X9C0 (Q7X9C0) NIN-like protein 2, complete 416 e-117
BI417701 118 4e-27
BP067107 79 3e-15
TC19287 42 4e-04
TC12819 37 0.011
TC18066 33 0.26
TC16422 similar to PIR|T05653|T05653 amino acid transport protei... 31 0.76
BP042603 31 0.99
TC16108 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide r... 30 2.2
TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete 29 3.8
BI418308 28 4.9
TC18053 weakly similar to GB|AAH54244.1|32484237|BC054244 MGC644... 28 4.9
TC9396 similar to UP|Q9ZRB9 (Q9ZRB9) Homeobox 1 protein (Fragmen... 28 4.9
TC12847 similar to GB|AAM66074.1|21595125|AY088542 endosperm-spe... 28 6.4
AV426846 28 8.4
AV772572 28 8.4
AV425721 28 8.4
>TC16033 UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (88%)
Length = 2554
Score = 1599 bits (4140), Expect = 0.0
Identities = 795/799 (99%), Positives = 797/799 (99%), Gaps = 1/799 (0%)
Frame = +3
Query: 106 GRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDML 165
GRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDML
Sbjct: 3 GRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDML 182
Query: 166 IQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRV 225
IQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRV
Sbjct: 183 IQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRV 362
Query: 226 FRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQI 285
FRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQI
Sbjct: 363 FRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQI 542
Query: 286 NYVPQLESVCKALEVVDLTSLKHSSIQNAK-ARDKSYEAALPEIQEVLRSACHMHKLPLA 344
NYVPQLESVCKALEVVDLTSLKHSSIQNAK ARDKSYEAALPEIQEVLRSACHMHKLPLA
Sbjct: 543 NYVPQLESVCKALEVVDLTSLKHSSIQNAKQARDKSYEAALPEIQEVLRSACHMHKLPLA 722
Query: 345 QTWVSCFQQGKDGCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGK 404
QTWVSCFQQGKDGCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGK
Sbjct: 723 QTWVSCFQQGKDGCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGK 902
Query: 405 AFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNC 464
AFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNC
Sbjct: 903 AFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNC 1082
Query: 465 NDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQH 524
NDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQH
Sbjct: 1083NDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQH 1262
Query: 525 ITSVASLEPEEKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTG 584
ITSVASLEPEEKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTG
Sbjct: 1263ITSVASLEPEEKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTG 1442
Query: 585 EKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGH 644
EKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGH
Sbjct: 1443EKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGH 1622
Query: 645 SLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPD 704
SLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPD
Sbjct: 1623SLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPD 1802
Query: 705 LGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRI 764
LGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRI
Sbjct: 1803LGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRI 1982
Query: 765 RSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGD 824
RSEAELKS SENKA+LFPRSLSQETLGEHTKTEYQSYLLKTCH+ATPKEDAHRVKVTYGD
Sbjct: 1983RSEAELKSPSENKAQLFPRSLSQETLGEHTKTEYQSYLLKTCHQATPKEDAHRVKVTYGD 2162
Query: 825 EKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCL 884
EKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCL
Sbjct: 2163EKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCL 2342
Query: 885 SSESSTIKLCIQASSGMRS 903
SSESSTIKLCIQASSGMRS
Sbjct: 2343SSESSTIKLCIQASSGMRS 2399
>TC14500 UP|Q9S7B1 (Q9S7B1) Nodule INCEPTION protein, complete
Length = 3001
Score = 521 bits (1342), Expect = e-148
Identities = 343/837 (40%), Positives = 467/837 (54%), Gaps = 66/837 (7%)
Frame = +3
Query: 128 KRWWIAPTCSPG--LGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRG-------DRP 178
KRWWI P + S+ E+L+ A+ ++K + N ++LIQIWVP+ RG
Sbjct: 345 KRWWIGPAAAVAGSCNSSVKERLVIAVGYLKDYTRNSNVLIQIWVPLRRGILHDHDYHTN 524
Query: 179 ILSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVR 238
L +NN P E A + +S GF +P P V VR
Sbjct: 525 YLLSNNPP----PQPEAAADHESVSLGFP----------MPAAPNSNLYSNV-----HVR 647
Query: 239 FFRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQ-INYVPQLESVCKA 297
FFRS EYPRV+ A+++ G+LA+P+FE+G+ TCLGV+E+V+T Q INY +V A
Sbjct: 648 FFRSHEYPRVQ-AQQY---GSLALPVFERGTGTCLGVLEIVITNQTTINY-----NVSNA 800
Query: 298 LE-VVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKD 356
L+ VD S + K D+ Y+AA+ EI EV+ S C H LPLA TW C QQGK
Sbjct: 801 LDQAVDFRSSQSFIPPAIKVYDELYQAAVNEIIEVMTSVCKTHNLPLALTWAPCIQQGKC 980
Query: 357 GCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTD 416
GC S +NY+ C+S V+ AC+VGD + F EAC E+HL +GQG+ G AF ++P F+ D
Sbjct: 981 GCGVSSENYMWCVSTVDSACFVGDLDILGFQEACSEYHLFRGQGIVGTAFTTSKPCFAID 1160
Query: 417 ITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYS-SADDYVLEFFLPVNCNDSEEQKNMLI 475
IT SK +YPL+HHA +FGL AAVAI LRS+Y+ SA D+VLEFFLP +C+DSEEQK +L
Sbjct: 1161 ITAFSKAEYPLAHHANMFGLHAAVAIPLRSVYTGSAADFVLEFFLPKDCHDSEEQKQLLN 1340
Query: 476 SLSIIIQRCCRSLRVI--------------TDKELER----------------TSSSVEV 505
SLS+++Q+ CRSL V+ + +ELE SS
Sbjct: 1341 SLSMVVQQACRSLHVVLVEDEYTLPMPSHTSKEELEEEEITITNNHEQKLFVSPSSHESE 1520
Query: 506 MALEDSGFARTVK--------------WSEPQH----ITSVASLEPEEKSSETVGGKFSD 547
+ E S A ++ EP+ T+ S ++ ++ F
Sbjct: 1521 CSKESSWIAHMMEAQQKGKGVSVSLEYLEEPKEEFKVTTNWDSSTDHDQQAQVFSSDFGQ 1700
Query: 548 LREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGS 607
+ + S ++G D+E S SS G K+GEKRR KA+KTI+L+VLRQYF GS
Sbjct: 1701 MSSGFKASTVEGG---DQESSYTFGSRRSSSGGRKSGEKRRTKAEKTISLQVLRQYFAGS 1871
Query: 608 LKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKID 667
LKDAAK+IGVC TTLKR+CRQHGI RWPSRKIKKVGHSL+KLQLVIDSVQGA GA +I
Sbjct: 1872 LKDAAKSIGVCPTTLKRICRQHGITRWPSRKIKKVGHSLKKLQLVIDSVQGAEGA-IQIG 2048
Query: 668 SFYSNFPDLASPNLSGA----SLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQ 723
SFY++FP+L+S + S + S N + N+L D G + + KS S+CSQ
Sbjct: 2049 SFYASFPELSSSDFSASCRSDSSKKMHNYPDQNNTLYGHGDHGGVVT--SLKSPPSACSQ 2222
Query: 724 GSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPR 783
+ C+ + + DV + P V + +L R + H E L
Sbjct: 2223 TFAGNQPCTII-----NNGDVLMTESPPV----PEALLSR-------RDHCEEAELLNNA 2354
Query: 784 SLSQETLG-EHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQ 842
S+ ++T K++ L + + + A RVK T+ DEK RF + W + L
Sbjct: 2355 SIQEDTKRFSRPKSQTLPPLSDSSGWNSLETGAFRVKATFADEKIRFSLQPIWGFSDLQL 2534
Query: 843 EIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI-QAS 898
EIARRFN++D++ +KYLDDD EWV+L CD DLEEC D+ SS+S TI+L + QAS
Sbjct: 2535 EIARRFNLNDVTNILLKYLDDDGEWVVLACDGDLEECKDIHRSSQSRTIRLSLFQAS 2705
>TC12706 UP|Q7X9C0 (Q7X9C0) NIN-like protein 2, complete
Length = 3814
Score = 416 bits (1069), Expect = e-117
Identities = 296/813 (36%), Positives = 428/813 (52%), Gaps = 65/813 (7%)
Frame = +3
Query: 135 TCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSE 194
T S GPS+ E+++RAL + K+ + +L Q+WVP+ G + ILS + P+ LD +
Sbjct: 828 TISRPPGPSLDERMLRALSFFKE-SAGGGILAQVWVPLEHGGQVILSTSEQPYLLD---Q 995
Query: 195 NLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREF 254
LA YRE+S F+F AE GLPGRVF KVPEWT +V ++ +EY RVEHAR +
Sbjct: 996 MLAGYREVSRTFKFPAEGKPGGF-SGLPGRVFVSKVPEWTSNVGYYSKNEYLRVEHARNY 1172
Query: 255 DICGTLAVPIFEQGSRT-CLGVIEVVMTTQQINYVPQLESVCKALEVVDL-TSLKHSSIQ 312
+ GT+A PIF+ S C V+E+V T + ++ +LE VC AL++V+L T++
Sbjct: 1173 KVRGTIAFPIFDTHSELPCCAVLELVTTKEMSDFDRELEVVCHALQLVNLRTTMPLRIFP 1352
Query: 313 NAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSC-FQQG----------KDGCRHS 361
+ +K AAL EI +VL+S CH H+LPLA TW+ C + +G K+G S
Sbjct: 1353 ECYSNNK--RAALAEIVDVLKSVCHAHRLPLALTWIPCCYTEGPKGEAMRIQIKEGHSSS 1526
Query: 362 EDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLS 421
+ L CI E ACYV D + F AC+EH L +G+G+AGKA N PFF D+
Sbjct: 1527 GEKVLLCIE--ESACYVTDRLMEGFVRACIEHPLEEGKGIAGKALQSNHPFFYPDVKEYD 1700
Query: 422 KTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIII 481
++YPL HHAR L AAVAIRLRS +++ DDY+LEFFLPVN S EQ+ +L +LS +
Sbjct: 1701 ISEYPLVHHARKCNLSAAVAIRLRSTHTNDDDYILEFFLPVNMRGSSEQQLLLDNLSGTM 1880
Query: 482 QRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETV 541
QR CRSLR ++D E R S+ GF E +++ S + L E +
Sbjct: 1881 QRICRSLRTVSDVESSRIEST-------HMGF-------ENKNLPSFSPLSRENSQIPLI 2018
Query: 542 GGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLR 601
+++ + S L+ P+ N S G + EK R A+K ++L VL+
Sbjct: 2019 NANQDSVQKSLKASRLRNK----GSKPPY---NQVSNGSRRQVEKNRGTAEKNVSLSVLQ 2177
Query: 602 QYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASG 661
Q+F GSLKDAAK+IGVC TTLKR+CR HGI RWPSRKI KV SL+K+Q V+DSVQG
Sbjct: 2178 QHFSGSLKDAAKSIGVCPTTLKRICRHHGISRWPSRKINKVNSSLKKIQTVLDSVQGVE- 2354
Query: 662 ASFKID----SFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSP------- 710
+S K D +F + + + + L + +P ++ P P
Sbjct: 2355 SSLKFDPSVGAFVAGGSTIQGIDTHKSLLFPEKSTIRDPRPITQDAVSVPAVPCGESEKS 2534
Query: 711 ----EGATKSLSSS---CSQGS-------LSSHSCS------SMPEQQPHTSDVA----- 745
EG K ++S C + S SSHS S EQ SD+A
Sbjct: 2535 AIKLEGKLKKTNASLVDCCEDSKSMAMDDRSSHSSSLWTKAQGSSEQASLGSDLAKRDKW 2714
Query: 746 -CNKDPV---------VGKDSA----DVVLKRIRSEAELKSHSENKAKLFPRSLSQETLG 791
N D + VG+ S D + + ++ ++ + + L S +
Sbjct: 2715 VLNNDGLRVENLKCNTVGQSSGSFTDDEMGIDVDNDEVVELNHPTCSSLTGSSNGSSPMI 2894
Query: 792 EHTKTEYQSYLLKTCHKATPKEDAHR--VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFN 849
+ + QS+ ++ K+T + + VK TY + RF+ S L +E+A RF
Sbjct: 2895 HGSSSSSQSFENQSKGKSTTVDRGSKIIVKATYRKDIIRFKFDPSAGCFKLYEEVAARFK 3074
Query: 850 VSDMSKFDVKYLDDDLEWVLLTCDADLEECIDV 882
+ + F +KYLDD+ EWV+L D+DL+EC+D+
Sbjct: 3075 LQN-GTFQLKYLDDEEEWVMLVSDSDLQECVDI 3170
>BI417701
Length = 552
Score = 118 bits (296), Expect = 4e-27
Identities = 55/83 (66%), Positives = 67/83 (80%)
Frame = +2
Query: 579 GISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRK 638
G K EK+R+ +K ++L VL+QYF GSLKDAAK+IGVC TTLKR+CRQHGI RWPSRK
Sbjct: 11 GSRKQVEKKRSTVEKNVSLSVLQQYFSGSLKDAAKSIGVCPTTLKRICRQHGISRWPSRK 190
Query: 639 IKKVGHSLQKLQLVIDSVQGASG 661
I KV SL+K+Q+V+DSVQG G
Sbjct: 191 INKVNRSLKKIQIVLDSVQGMEG 259
>BP067107
Length = 541
Score = 79.0 bits (193), Expect = 3e-15
Identities = 44/98 (44%), Positives = 61/98 (61%)
Frame = +1
Query: 143 SIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREI 202
S+ E++++AL + K+ + +L Q+WVPV GD ILS + P+ LD LA YRE+
Sbjct: 253 SLDERMLKALSFFKE-SAGGGILAQVWVPVKHGDEFILSTSEQPYLLD---HKLAGYREV 420
Query: 203 SEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFF 240
S F FSAE + PGLPGRVF VPEWT +V ++
Sbjct: 421 SRTFTFSAEGKAGSC-PGLPGRVFTSHVPEWTSNVGYY 531
>TC19287
Length = 603
Score = 42.0 bits (97), Expect = 4e-04
Identities = 18/41 (43%), Positives = 26/41 (62%)
Frame = +1
Query: 610 DAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQ 650
+A++ + V T LK+ CR+ GI RWP RKIK + + LQ
Sbjct: 13 EASRRLNVGLTVLKKKCREFGIPRWPHRKIKSLDSLIHDLQ 135
>TC12819
Length = 498
Score = 37.4 bits (85), Expect = 0.011
Identities = 21/34 (61%), Positives = 23/34 (66%)
Frame = -2
Query: 178 PILSANNLPFSLDSGSENLARYREISEGFQFSAE 211
PILSANNLPF LDSG ++ E S G QFS E
Sbjct: 392 PILSANNLPFLLDSGLQS----TETSVGCQFSTE 303
>TC18066
Length = 480
Score = 32.7 bits (73), Expect = 0.26
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Frame = -2
Query: 765 RSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPK-EDAHRVKVTYG 823
R A+ K S ++ LFP S TL ++++LL TC T K EDA + G
Sbjct: 305 RYTAKQKKWSR*ESFLFPFSSDVLTLYNSNSLYFRNWLLVTCSTCTVKVEDAEPLITDEG 126
Query: 824 DEKTRFRMPKSWSYE 838
E+ R R+ + E
Sbjct: 125 GERQRLRLADNGGLE 81
>TC16422 similar to PIR|T05653|T05653 amino acid transport protein homolog
F22I13.20 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (38%)
Length = 685
Score = 31.2 bits (69), Expect = 0.76
Identities = 34/140 (24%), Positives = 48/140 (34%), Gaps = 2/140 (1%)
Frame = +3
Query: 674 PDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSS 733
P + P L L S PNS P S ++ SL++S + SCSS
Sbjct: 120 PTPSHPTLEKTHLSSPNPHLSPPNSKPSPTSSSPSSAPESSASLTASSEPAGSPASSCSS 299
Query: 734 MPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRS--LSQETLG 791
P T+ C+ P+ + A S S ++ P + LS L
Sbjct: 300 PSPSSPTTA--*CSSSPLAASSIPSLGTPNSDPSATSASPSAARSAASPSTP*LSSPRLD 473
Query: 792 EHTKTEYQSYLLKTCHKATP 811
+ T S L ATP
Sbjct: 474 SASATSSSSRPLSASSPATP 533
>BP042603
Length = 565
Score = 30.8 bits (68), Expect = 0.99
Identities = 16/43 (37%), Positives = 23/43 (53%)
Frame = +3
Query: 546 SDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRR 588
S+L HQ+D + GN+ D S GN V ++TG KR+
Sbjct: 156 SNLNMHQQDKVAGGNVNVDGSMSNTFRGN-DQVTKNQTGRKRK 281
>TC16108 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide
repeat(TPR)-containing protein-like, partial (3%)
Length = 597
Score = 29.6 bits (65), Expect = 2.2
Identities = 14/47 (29%), Positives = 29/47 (60%)
Frame = +3
Query: 649 LQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSEN 695
L ++SVQ S K+ + + ++PD+AS + S+++AL+ S++
Sbjct: 69 LNSFMNSVQSWLYPSLKVTTSWKDYPDIASTFTTTGSVIAALSSSDD 209
>TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete
Length = 1064
Score = 28.9 bits (63), Expect = 3.8
Identities = 41/143 (28%), Positives = 57/143 (39%), Gaps = 19/143 (13%)
Frame = -2
Query: 663 SFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLS---PEGATKSL-- 717
SF + S + F LASP LN + + + L D G S P ++ S
Sbjct: 802 SFLMVSLMNRFSALASPR------AWFLNTTSSSHLLFTLKDAGLFSNGFPRRSSASFTL 641
Query: 718 -SSSCSQGSLSSHS-------CSS------MPEQQPHTSDVACNKDPVVGKDSADVVLKR 763
SSSCS S S S CSS + +S ++ + + SA +
Sbjct: 640 SSSSCSSRSFRSFSSALSFFICSSSASKASVSSSSSSSSSLSSSLLTSTSESSASTLRAT 461
Query: 764 IRSEAELKSHSENKAKLFPRSLS 786
I S L S+N+ LFP SLS
Sbjct: 460 IWSSTSLFVPSKNRWLLFP*SLS 392
>BI418308
Length = 411
Score = 28.5 bits (62), Expect = 4.9
Identities = 12/45 (26%), Positives = 20/45 (43%)
Frame = +2
Query: 169 WVPVPRGDRPILSANNLPFSLDSGSENLARYREISEGFQFSAEED 213
W P P D P++ N PF + G L+ ++ F+ +D
Sbjct: 59 WPPAPLPDTPLVIVANHPFGIGDGIAVLSLVEQLGRPFRVMIHKD 193
>TC18053 weakly similar to GB|AAH54244.1|32484237|BC054244 MGC64450 protein
{Xenopus laevis;} , partial (20%)
Length = 704
Score = 28.5 bits (62), Expect = 4.9
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 2/37 (5%)
Frame = -1
Query: 857 DVKYLDDDLEWVLLTCDADLEECIDVC--LSSESSTI 891
D +L D+EW+L+ C + EE + VC L S SST+
Sbjct: 176 DCVFLSLDMEWILI*C--EFEERVVVCFFLHSPSSTV 72
>TC9396 similar to UP|Q9ZRB9 (Q9ZRB9) Homeobox 1 protein (Fragment),
partial (68%)
Length = 1138
Score = 28.5 bits (62), Expect = 4.9
Identities = 15/47 (31%), Positives = 24/47 (50%), Gaps = 8/47 (17%)
Frame = +1
Query: 531 LEPEEKSSETVGGKFSDLREHQED--------SILKGNIECDRECSP 569
L+PE ++E GG DLRE++ D +L ++ C R +P
Sbjct: 46 LQPETSTAEKSGGGGEDLREYKADILGHQLYEQLLSAHVSCLRIATP 186
>TC12847 similar to GB|AAM66074.1|21595125|AY088542 endosperm-specific
protein-like protein {Arabidopsis thaliana;} , partial
(11%)
Length = 245
Score = 28.1 bits (61), Expect = 6.4
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = +1
Query: 698 SLSIQPDLGPLSPEGATKSLSSS 720
SLS+ P L P+SP+ T LSSS
Sbjct: 10 SLSLPPPLSPISPKWVTTPLSSS 78
>AV426846
Length = 427
Score = 27.7 bits (60), Expect = 8.4
Identities = 13/38 (34%), Positives = 18/38 (47%)
Frame = +2
Query: 806 CHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQE 843
CH +T +KV YG+E R K W H ++E
Sbjct: 35 CHSSTGVFCRACLKVRYGEEIEEVRENKGWMCPHCIEE 148
>AV772572
Length = 491
Score = 27.7 bits (60), Expect = 8.4
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = -3
Query: 610 DAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKK 641
DA++ +G T K R+HG K+W K K
Sbjct: 294 DASRALGDLTGIKKHFSRKHGEKKWKCEKCSK 199
>AV425721
Length = 392
Score = 27.7 bits (60), Expect = 8.4
Identities = 14/36 (38%), Positives = 18/36 (49%), Gaps = 3/36 (8%)
Frame = +2
Query: 381 PSVRFF---HEACMEHHLLKGQGVAGKAFMINQPFF 413
P +R F E HHLL +G + F IN+P F
Sbjct: 47 PKMRLFGFPEEILHHHHLLHSEGCS*ARFEINEPLF 154
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,296,092
Number of Sequences: 28460
Number of extensions: 239053
Number of successful extensions: 1364
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1329
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1344
length of query: 903
length of database: 4,897,600
effective HSP length: 99
effective length of query: 804
effective length of database: 2,080,060
effective search space: 1672368240
effective search space used: 1672368240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0193.5


