

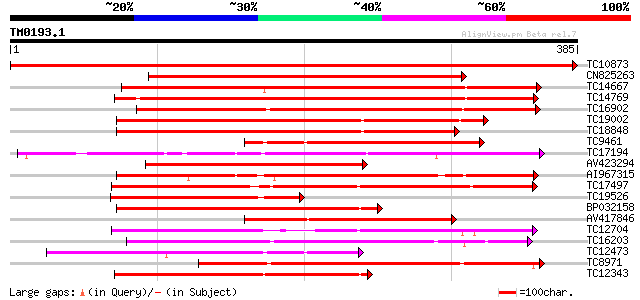
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.1
(385 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 773 0.0
CN825263 251 1e-67
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 240 3e-64
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 233 5e-62
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 228 1e-60
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 220 3e-58
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 217 3e-57
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 210 4e-55
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 209 5e-55
AV423294 204 2e-53
AI967315 199 5e-52
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 196 4e-51
TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like ... 190 4e-49
BP032158 189 7e-49
AV417846 187 3e-48
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 183 4e-47
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 182 1e-46
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 177 2e-45
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 171 2e-43
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 167 4e-42
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 773 bits (1997), Expect = 0.0
Identities = 384/385 (99%), Positives = 384/385 (99%)
Frame = +1
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS
Sbjct: 199 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 378
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR 120
NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR
Sbjct: 379 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR 558
Query: 121 QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL 180
QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL
Sbjct: 559 QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL 738
Query: 181 NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 240
NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD
Sbjct: 739 NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 918
Query: 241 NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 300
NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV
Sbjct: 919 NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 1098
Query: 301 SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 360
SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY
Sbjct: 1099SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 1278
Query: 361 LASQGSPEAHYLYGVQQRPSRMERN 385
LASQGSPEAHYLYGVQQ PSRMERN
Sbjct: 1279LASQGSPEAHYLYGVQQPPSRMERN 1353
>CN825263
Length = 663
Score = 251 bits (642), Expect = 1e-67
Identities = 123/216 (56%), Positives = 159/216 (72%)
Frame = +1
Query: 95 GFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQR 154
GFG VYKG L G VAVK L D ++G +EF+ EV MLS LHH NLV+LIG C + R
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 155 LLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKS 214
L+YE +P GS+E HL + PL+W+ RMK+A+GAARGL YLH ++P VI+RD KS
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 215 ANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSF 274
+NILL+ +F PK+SDFGLA+ N H+ST VMGT+GY APEYAM+G L +KSD+YS+
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSY 540
Query: 275 GVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDR 310
GVVLLELLTG + +D S+ PG++NLV+WARP + +
Sbjct: 541 GVVLLELLTGTKPVDLSQPPGQENLVTWARPILTSK 648
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 240 bits (612), Expect = 3e-64
Identities = 130/291 (44%), Positives = 188/291 (63%), Gaps = 6/291 (2%)
Frame = +2
Query: 77 ELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR-QGFQEFVMEVLMLSL 135
EL + T NF LIGEG +G+VY L G AVAVK+L + EF+ +V M+S
Sbjct: 335 ELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSR 514
Query: 136 LHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLF-----ELSHDKEPLNWSTRMKVAV 190
L + N V L GYC +G+ R+L YE+ MGSL D L + + L+W R+++AV
Sbjct: 515 LKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAV 694
Query: 191 GAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMG 250
AARGLEYLH P +I+RD++S+N+L+ ++ K++DF L+ P STRV+G
Sbjct: 695 DAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLG 874
Query: 251 TYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDR 310
T+GY APEYAM+G+LT KSD+YSFGVVLLELLTGR+ +D + G+Q+LV+WA P S+
Sbjct: 875 TFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSE- 1051
Query: 311 RRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
+ VDP L+G +P + + + A+ A+C+Q + +FRP ++ +V AL+ L
Sbjct: 1052DKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPL 1204
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 233 bits (593), Expect = 5e-62
Identities = 123/288 (42%), Positives = 179/288 (61%)
Frame = +3
Query: 72 SFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVL 131
+F + AT +K LIGEGGFG VY+G L G+ VAVK S QG +EF E+
Sbjct: 2130 AFTLEYIEVATERYK--TLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELN 2303
Query: 132 MLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVG 191
+LS + H NLV L+GYC + DQ++LVY +M GSL+D L+ ++ L+W TR+ +A+G
Sbjct: 2304 LLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALG 2483
Query: 192 AARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGT 251
AARGL YLH VI+RD+KS+NILLD+ K++DFG +K P +++VS V GT
Sbjct: 2484 AARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGT 2663
Query: 252 YGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRR 311
GY PEY + +L+ KSD++SFGVVLLE+++GR ++ R E +LV WA PY
Sbjct: 2664 AGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRG-S 2840
Query: 312 RFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
+ +VDP ++G + + + + + + CL+ +RP + IV LE
Sbjct: 2841 KVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELE 2984
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 228 bits (581), Expect = 1e-60
Identities = 120/274 (43%), Positives = 171/274 (61%)
Frame = +3
Query: 87 EANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIG 146
EA L+G GGFGKVY G + G VA+K+ + QG EF E+ MLS L H +LV LIG
Sbjct: 3 EALLLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIG 182
Query: 147 YCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPP 206
YC + + +LVY++M G+L +HL++ K PL W R+++ +GAARGL YLH A
Sbjct: 183 YCEENTEMILVYDHMAYGTLREHLYKTQ--KPPLPWKQRLEICIGAARGLHYLHTGAKYT 356
Query: 207 VIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLT 266
+I+RD+K+ NILLD ++ K+SDFGL+K GP DNTHVST V G++GY PEY +LT
Sbjct: 357 IIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLT 536
Query: 267 LKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFP 326
KSD+YSFGVVL E+L R A++ S + +L WA + ++ ++DP L+G+
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCY-NKGILDQILDPYLKGKIA 713
Query: 327 SRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 360
C + C+ +Q RP + D++ LE+
Sbjct: 714 PECFKKFAETAMKCVSDQGIERPSMGDVLWNLEF 815
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 220 bits (561), Expect = 3e-58
Identities = 112/253 (44%), Positives = 163/253 (64%)
Frame = +1
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLM 132
F +EL AT NF N +GEGGFG VY G+L G +AVK+L + EF +EV +
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEI 207
Query: 133 LSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGA 192
L+ + H NL+ L GYC +G +RL+VY+YMP SL HL + L+W+ RM +A+G+
Sbjct: 208 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS 387
Query: 193 ARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTY 252
A G+ YLH A P +I+RD+K++N+LLD++F +++DFG AKL P G THV+TRV GT
Sbjct: 388 AEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDG-ATHVTTRVKGTL 564
Query: 253 GYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRR 312
GY APEYAM GK D++SFG++LLEL +G++ ++ ++++ WA P + ++
Sbjct: 565 GYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALP-LACAKK 741
Query: 313 FGHMVDPLLQGRF 325
F DP L G +
Sbjct: 742 FTEFADPRLNGEY 780
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 217 bits (552), Expect = 3e-57
Identities = 110/233 (47%), Positives = 154/233 (65%)
Frame = +2
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLM 132
F ++EL AT F + N +GEGGFG VY GR + G +AVK+L + EF +EV +
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 445
Query: 133 LSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGA 192
L + H NL+ L GYC DQRL+VY+YMP SL HL + LNW RMK+A+G+
Sbjct: 446 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGS 625
Query: 193 ARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTY 252
A G+ YLH P +I+RD+K++N+LL+++F P ++DFG AKL P G +H++TRV GT
Sbjct: 626 AEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEG-VSHMTTRVKGTL 802
Query: 253 GYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARP 305
GY APEYAM GK++ D+YSFG++LLEL+TGR+ I+ ++ + WA P
Sbjct: 803 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGVKRTITEWAEP 961
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 210 bits (534), Expect = 4e-55
Identities = 105/163 (64%), Positives = 125/163 (76%)
Frame = +3
Query: 160 YMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILL 219
+MP GSLE+HLF +PL+WS RMKVA+GAARGL +LH A VIYRD K++NILL
Sbjct: 3 FMPKGSLENHLFR--RGPQPLSWSVRMKVAIGAARGLSFLH-NAKSQVIYRDFKASNILL 173
Query: 220 DNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLL 279
D EFN KLSDFGLAK GP GD THVST+VMGT GY APEY +G+LT KSD+YSFGVVLL
Sbjct: 174 DAEFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLL 353
Query: 280 ELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQ 322
ELL+GRRA+D + +QNLV WARPY D+RR ++D L+
Sbjct: 354 ELLSGRRAVDKTIAGVDQNLVDWARPYLGDKRRLFRIMDSKLE 482
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 209 bits (533), Expect = 5e-55
Identities = 134/366 (36%), Positives = 205/366 (55%), Gaps = 8/366 (2%)
Frame = +1
Query: 6 CLFSR--RKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNGS 63
C++ R +K+ + ++ + + + S+ ++A T G G G +S+ G
Sbjct: 862 CMYVRYQKKEEEKAKLPTDISMALSTQDGNASSSAEYETSGS-------SGPGTASATGL 1020
Query: 64 SNGKTAAA-SFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQG 122
++ A + F ++ELA AT NF N IG+GGFG VY L G+ A+K++ Q
Sbjct: 1021 TSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDV---QA 1188
Query: 123 FQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNW 182
EF+ E+ +L+ +HH NLVRLIGYC +G LVYE++ G+L +L KEPL W
Sbjct: 1189 STEFLCELKVLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLH--GSGKEPLPW 1359
Query: 183 STRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNT 242
S+R+++A+ AARGLEY+H P I+RD+KSANIL+D K++DFGL KL VG++T
Sbjct: 1360 SSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNST 1539
Query: 243 HVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAI-----DTSRRPGEQ 297
+ TR++GT+GY PEYA G ++ K D+Y+FGVVL EL++ + A+ + G
Sbjct: 1540 -LQTRLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLV 1716
Query: 298 NLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVA 357
L A +VDP L +P + + + C ++ P RP + +VVA
Sbjct: 1717 ALFEEALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVA 1896
Query: 358 LEYLAS 363
L L+S
Sbjct: 1897 LMTLSS 1914
>AV423294
Length = 459
Score = 204 bits (520), Expect = 2e-53
Identities = 96/152 (63%), Positives = 123/152 (80%), Gaps = 1/152 (0%)
Frame = +3
Query: 93 EGGFGKVYKGRLTT-GEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDG 151
EGGFGKVY+G + + VA+KQL+ G QG +EFV+EVL LSL H NLV+LIG+C +G
Sbjct: 3 EGGFGKVYRGFIERINQVVAIKQLNPHGLQGIREFVVEVLTLSLADHPNLVKLIGFCAEG 182
Query: 152 DQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRD 211
+QRLLVYEYMP+GSLE HL ++ K+PL+W+TRMK+A GAARGLEYLH PPVIYRD
Sbjct: 183 EQRLLVYEYMPLGSLESHLHDILPGKKPLDWNTRMKIAAGAARGLEYLHNKMKPPVIYRD 362
Query: 212 LKSANILLDNEFNPKLSDFGLAKLGPVGDNTH 243
+K +N+L+ + ++PKLSDFGLAK+GP+GD TH
Sbjct: 363 IKCSNLLIGDGYHPKLSDFGLAKVGPIGDKTH 458
>AI967315
Length = 1308
Score = 199 bits (507), Expect = 5e-52
Identities = 108/291 (37%), Positives = 178/291 (61%), Gaps = 4/291 (1%)
Frame = +1
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR--QGFQEFVMEV 130
F + EL AT F N++G+GG+ +VYKGRL +G+ +AVK+L+ R + +EF+ E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 131 LMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKE--PLNWSTRMKV 188
+ + H+N++ L+G C D LV+E +GS+ L HD++ PL+W TR K+
Sbjct: 292 GTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVAS----LIHDEKMAPLDWKTRYKI 456
Query: 189 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRV 248
+G ARGL YLH +I+RD+K++NILL +F P++SDFGLAK P H +
Sbjct: 457 VLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPI 636
Query: 249 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFS 308
GT+G+ APEY M G + K+D+++FGV LLE+++GR+ +D S Q+L +WA+P S
Sbjct: 637 EGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS----HQSLHTWAKPILS 804
Query: 309 DRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
+ +VDP L+G + ++ ++C++ +RP +++++ +E
Sbjct: 805 -KWEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVME 954
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 196 bits (499), Expect = 4e-51
Identities = 114/290 (39%), Positives = 175/290 (60%), Gaps = 1/290 (0%)
Frame = +1
Query: 70 AASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVME 129
A F F ++ AT +F +N +GEGGFG VYKG L G+ +AVK+LS+ QG +EF E
Sbjct: 1627 ATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNE 1806
Query: 130 VLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVA 189
+ +++ L H NLV+L G D+ + M + L + + K ++W+ R+++
Sbjct: 1807 IKLIARLQHRNLVKLFGCSVHQDENSHANKKMKI------LLDSTRSK-LVDWNKRLQII 1965
Query: 190 VGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVST-RV 248
G ARGL YLH + +I+RDLK++NILLD+E NPK+SDFGLA++ +GD T RV
Sbjct: 1966 DGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIF-IGDQVEARTKRV 2142
Query: 249 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFS 308
MGTYGY PEYA+ G ++KSD++SFGV++LE+++G++ NL+S A +
Sbjct: 2143 MGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWI 2322
Query: 309 DRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVAL 358
+ R +VD LL + + I + +C+Q +P+ RP + IV+ L
Sbjct: 2323 EERPL-ELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLML 2469
>TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (37%)
Length = 432
Score = 190 bits (482), Expect = 4e-49
Identities = 95/133 (71%), Positives = 112/133 (83%), Gaps = 1/133 (0%)
Frame = +1
Query: 69 AAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGRQGFQEFV 127
AA +F FRELA AT+NF+ L+GEGGFG+VYKGRL +TG+ VAVKQL +G QG +EF+
Sbjct: 40 AAQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLESTGQVVAVKQLDRNGLQGNREFL 219
Query: 128 MEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMK 187
+EVLMLSLLHH NLV LIGYC DGDQRLLVYE+MP+GSLEDH L DKEPL+W+TRMK
Sbjct: 220 VEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDH---LHGDKEPLDWNTRMK 390
Query: 188 VAVGAARGLEYLH 200
+A GAA+GLEYLH
Sbjct: 391 IAAGAAKGLEYLH 429
>BP032158
Length = 555
Score = 189 bits (480), Expect = 7e-49
Identities = 96/181 (53%), Positives = 123/181 (67%)
Frame = +2
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLM 132
F R++ AT NF AN IGEGGFG VYKG L+ G+ +AVKQLS +QG +EF+ E+ M
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFINEIGM 193
Query: 133 LSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGA 192
+S L H NLV+L G C +G+Q LLVYEYM SL LF K LNW TRMK+ VG
Sbjct: 194 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVGI 373
Query: 193 ARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTY 252
A+GL YLH + +++RD+K+ N+LLD + K+SDFGLAKL +NTH+STR+ GT
Sbjct: 374 AKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDE-EENTHISTRIAGTI 550
Query: 253 G 253
G
Sbjct: 551 G 553
>AV417846
Length = 429
Score = 187 bits (475), Expect = 3e-48
Identities = 89/144 (61%), Positives = 114/144 (78%)
Frame = +1
Query: 160 YMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILL 219
+MP GS+E+HLF +P +WS R+K+A+GAA+GL +LH T +P VIYRD K++NILL
Sbjct: 1 FMPKGSMENHLFRRGSYFQPFSWSLRLKIALGAAKGLAFLHST-EPKVIYRDFKTSNILL 177
Query: 220 DNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLL 279
D +++ KLSDFGLA+ GPVGD +HVSTRVMGT GY APEY +G LT SD+YSFGVVLL
Sbjct: 178 DTKYSAKLSDFGLARDGPVGDKSHVSTRVMGTRGYAAPEYLATGHLTANSDVYSFGVVLL 357
Query: 280 ELLTGRRAIDTSRRPGEQNLVSWA 303
E+++GRRAID ++ GE NLV WA
Sbjct: 358 EIISGRRAIDKNQLAGEHNLVEWA 429
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 183 bits (465), Expect = 4e-47
Identities = 111/313 (35%), Positives = 175/313 (55%), Gaps = 24/313 (7%)
Frame = +1
Query: 70 AASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVME 129
A F F ++ T +F E+N +GEGGFG VYKG L G+ +AVK+LS+ QG +EF E
Sbjct: 1408 ATIFDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTSGQGMEEFKNE 1587
Query: 130 VLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVA 189
V +++ L H NLV+L+G D+ LL+YE+M SL+ +F+ +R+++
Sbjct: 1588 VKLIARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIFD-----------SRLRI- 1731
Query: 190 VGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVST-RV 248
I+RDLK++NILLD+E NPK+SDFGLA++ GD T RV
Sbjct: 1732 ------------------IHRDLKTSNILLDSEMNPKISDFGLARIF-TGDQVEAKTKRV 1854
Query: 249 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPY-- 306
MGTYGY +PEYA+ G ++KSD++SFGV++LE+++G++ +NL+S + +
Sbjct: 1855 MGTYGYMSPEYAVHGSFSVKSDVFSFGVIVLEIISGKKIGRFCDPHHHRNLLSHSSNFAV 2034
Query: 307 -------------FSDRRRFG--------HMVDPLLQGRFPSRCLHQAIAITAMCLQEQP 345
+R+ + +VD LL G + + I I +C+Q++P
Sbjct: 2035 FLIKALRICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTEILRYIHIALLCVQQRP 2214
Query: 346 KFRPLITDIVVAL 358
++RP + +V+ L
Sbjct: 2215 EYRPDMLSVVLML 2253
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 182 bits (461), Expect = 1e-46
Identities = 112/283 (39%), Positives = 156/283 (54%), Gaps = 7/283 (2%)
Frame = +2
Query: 80 DATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR-QGFQEFVMEVLMLSLLHH 138
D KE N+IG+GG G VY+G + G VA+K+L G + F E+ L + H
Sbjct: 2177 DVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRH 2356
Query: 139 TNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEY 198
N++RL+GY ++ D LL+YEYMP GSL + L L W R K+AV AARGL Y
Sbjct: 2357 RNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH--LRWEMRYKIAVEAARGLCY 2530
Query: 199 LHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPE 258
+H P +I+RD+KS NILLD +F ++DFGLAK + + + G+YGY APE
Sbjct: 2531 MHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPE 2710
Query: 259 YAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYF------SDRRR 312
YA + K+ KSD+YSFGVVLLEL+ GR+ + ++V W SD
Sbjct: 2711 YAYTLKVDEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSELSQPSDTAL 2884
Query: 313 FGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIV 355
+VDP L G +P + I MC++E RP + ++V
Sbjct: 2885 VLAVVDPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVV 3010
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 177 bits (450), Expect = 2e-45
Identities = 98/225 (43%), Positives = 136/225 (59%), Gaps = 10/225 (4%)
Frame = +3
Query: 26 SGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNGSSNGKTAAASFGFRELADATRNF 85
+G +S G ++ + + + E+ V +G F F +L AT++F
Sbjct: 279 TGRTSSGSGKSSMSQVWEINTESESGSVVNNVKGGSGEFLEVAKLKVFSFGDLKSATKSF 458
Query: 86 KEANLIGEGGFGKVYKGRLT----------TGEAVAVKQLSHDGRQGFQEFVMEVLMLSL 135
K LIGEGGFGKVYKG L +G VAVK+L+ + QGF E+ E+ L
Sbjct: 459 KADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPESMQGFHEWQSEINFLGR 638
Query: 136 LHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARG 195
+ H NLV+L+GYC D ++ LLVYE+MP GSLE+HLF + E L+W+TR+K+A+GAARG
Sbjct: 639 ISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFR--RNTESLSWNTRLKIAIGAARG 812
Query: 196 LEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 240
L +LH + VIYRD K++NILLD +N K+S FGL K GP G+
Sbjct: 813 LAFLH-SLXKIVIYRDFKASNILLDGNYNAKISXFGLTKFGPSGE 944
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 171 bits (433), Expect = 2e-43
Identities = 94/242 (38%), Positives = 150/242 (61%), Gaps = 7/242 (2%)
Frame = +3
Query: 129 EVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKV 188
EV +L+ + H NLV+L+G+ G++R+L+ EY+P G+L +HL L + L+++ R+++
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRG--KILDFNQRLEI 179
Query: 189 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPV-GDNTHVSTR 247
A+ A GL YLH A+ +I+RD+KS+NILL K++DFG A+LGPV GD TH+ST+
Sbjct: 180 AIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK 359
Query: 248 VMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWA-RPY 306
V GT GY PEY + +LT KSD+YSFG++LLE+LTGRR ++ + E+ + WA R Y
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 307 FSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDI-----VVALEYL 361
+ ++DPL++ + L + + ++ C RP + + + +YL
Sbjct: 540 --NEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQLWAIRADYL 713
Query: 362 AS 363
S
Sbjct: 714 KS 719
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 167 bits (422), Expect = 4e-42
Identities = 84/175 (48%), Positives = 116/175 (66%)
Frame = +3
Query: 72 SFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVL 131
+F + L AT+NF N +GEGGFG V+KG+L G +AVK+LS QG +F+ E
Sbjct: 204 TFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQGRTQFINEAK 383
Query: 132 MLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVG 191
+L+ + H N+V L GYC G ++LLVYEY+P SL+ LF S KE L+W R + G
Sbjct: 384 LLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFR-SQKKEQLDWKRRFDIISG 560
Query: 192 AARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVST 246
ARGL YLH + +I+RD+K+ANILLD ++ PK++DFGLA++ P D THV+T
Sbjct: 561 VARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFP-EDQTHVNT 722
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,295,784
Number of Sequences: 28460
Number of extensions: 82977
Number of successful extensions: 932
Number of sequences better than 10.0: 333
Number of HSP's better than 10.0 without gapping: 746
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 755
length of query: 385
length of database: 4,897,600
effective HSP length: 92
effective length of query: 293
effective length of database: 2,279,280
effective search space: 667829040
effective search space used: 667829040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0193.1


