

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0189.12
(963 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
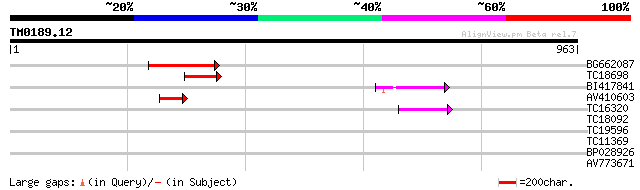
Score E
Sequences producing significant alignments: (bits) Value
BG662087 213 2e-55
TC18698 75 5e-14
BI417841 59 4e-09
AV410603 52 4e-07
TC16320 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial ... 47 2e-05
TC18092 32 0.37
TC19596 31 0.82
TC11369 weakly similar to UP|BAC99695 (BAC99695) Brain protein 4... 28 5.3
BP028926 28 5.3
AV773671 28 6.9
>BG662087
Length = 373
Score = 213 bits (541), Expect = 2e-55
Identities = 102/120 (85%), Positives = 107/120 (89%)
Frame = +1
Query: 237 NGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDED 296
+GKWRM VD TDLNK CPKDSYPLPSIDKLVDGA NELLSLMDAYSGYHQIKMH SDED
Sbjct: 13 SGKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDED 192
Query: 297 KTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNH 356
KT FMTARVNYCYQT+PFGLKNAGATYQ LMDRVF VGRNMEVY+D+MIVKS L +NH
Sbjct: 193 KTAFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKSALRANH 372
>TC18698
Length = 808
Score = 75.1 bits (183), Expect = 5e-14
Identities = 38/64 (59%), Positives = 44/64 (68%)
Frame = -2
Query: 297 KTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNH 356
KTT RVNY YQ MP GLKN TYQRLMD++F Q+ +N+EVYV+DMIVKS H
Sbjct: 801 KTTLKINRVNYYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKSSQE*FH 622
Query: 357 HEDL 360
DL
Sbjct: 621 RGDL 610
>BI417841
Length = 617
Score = 58.9 bits (141), Expect = 4e-09
Identities = 41/130 (31%), Positives = 58/130 (44%), Gaps = 4/130 (3%)
Frame = +1
Query: 622 DGSSNDNGSEAG----VTLQGPGELVLEQSLRFQFKTSNNQAEYEALIAGLKLAIEVQID 677
DGSS N AG + + ++ L + + Q +NNQAEY LI GLK A E
Sbjct: 151 DGSSKGNPGSAGAGAVLRAEDGSKVYLREGVGNQ---TNNQAEYRGLILGLKHAHEQGYQ 321
Query: 678 SLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPRAQNQRADALA 737
+ V+ DS LV QV G ++ + + + L + Q + VPR N AD A
Sbjct: 322 HINVKGDSQLVCKQVEGSWKARNPNIASLCNEAKELKSKFQSFDINHVPRQYNSEADVQA 501
Query: 738 KLASTRKPGN 747
L G+
Sbjct: 502 NLGVNLPAGH 531
>AV410603
Length = 162
Score = 52.0 bits (123), Expect = 4e-07
Identities = 22/48 (45%), Positives = 32/48 (65%)
Frame = +1
Query: 255 KDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMT 302
KDS+P+P++D+L+D G++ S +D SGYHQI + D KT F T
Sbjct: 10 KDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRT 153
>TC16320 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (11%)
Length = 632
Score = 46.6 bits (109), Expect = 2e-05
Identities = 27/91 (29%), Positives = 40/91 (43%)
Frame = +2
Query: 661 YEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQV 720
Y LI GLK AI+ + V+ DS LV NQV G +++K + + L +
Sbjct: 2 YRGLILGLKHAIKEGYKHIQVKGDSMLVCNQVQGLWKIKNQNIASLCSEAKELKNKFLSF 181
Query: 721 VVEFVPRAQNQRADALAKLASTRKPGNNRSV 751
+ +PR N AD A + + G V
Sbjct: 182 KINHIPREYNSEADVQANFGISLRAGQVEEV 274
>TC18092
Length = 519
Score = 32.3 bits (72), Expect = 0.37
Identities = 18/35 (51%), Positives = 22/35 (62%)
Frame = -2
Query: 242 MCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELL 276
MCV TD+NK CP+ PL SI+ LVD + E L
Sbjct: 101 MCVC*TDINKDCPR--XPLTSINALVDYSSSYEYL 3
>TC19596
Length = 499
Score = 31.2 bits (69), Expect = 0.82
Identities = 27/86 (31%), Positives = 38/86 (43%), Gaps = 1/86 (1%)
Frame = +1
Query: 658 QAEYEALIAGLKLAIEVQIDSLPVRTDS-PLVANQVNGEFQVKELALIKYVERVRLLMGR 716
+AE AL GL L + D +V N +Q LA + +RLL+ R
Sbjct: 130 RAEIAALKHGLTLLWNAHVRRATCEVDCLDIVEALENDRYQFHALA--SELLDIRLLLDR 303
Query: 717 LQQVVVEFVPRAQNQRADALAKLAST 742
V + +VPR N AD LA L ++
Sbjct: 304 DWTVTLAYVPREANAAADCLAGLGAS 381
>TC11369 weakly similar to UP|BAC99695 (BAC99695) Brain protein 44-like,
partial (88%)
Length = 753
Score = 28.5 bits (62), Expect = 5.3
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Frame = -3
Query: 551 R*K*ELTFL*DRCCRSQTFQEGSS-----RDQWSCRNMDCSMTKGGRS 593
R *++ FL C +EGSS + +W DCS T GGRS
Sbjct: 337 RSN*QIWFLVVSLCPLLPQEEGSS*R*DPKSEWFLDQWDCSKTSGGRS 194
>BP028926
Length = 390
Score = 28.5 bits (62), Expect = 5.3
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = -3
Query: 866 KECAEFVRKCKKCQVFTDLPRAPP 889
+ C+ V C+KC+V++ LPR P
Sbjct: 103 ENCS*SVCNCQKCEVYSQLPRTKP 32
>AV773671
Length = 453
Score = 28.1 bits (61), Expect = 6.9
Identities = 12/24 (50%), Positives = 15/24 (62%)
Frame = +2
Query: 876 KKCQVFTDLPRAPPGQLVTISSPW 899
KKC FTD RAP G V++ P+
Sbjct: 254 KKCSFFTDCQRAPAGGDVSLPPPF 325
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,349,633
Number of Sequences: 28460
Number of extensions: 220293
Number of successful extensions: 841
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 840
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 841
length of query: 963
length of database: 4,897,600
effective HSP length: 99
effective length of query: 864
effective length of database: 2,080,060
effective search space: 1797171840
effective search space used: 1797171840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0189.12


