

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.7
(199 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
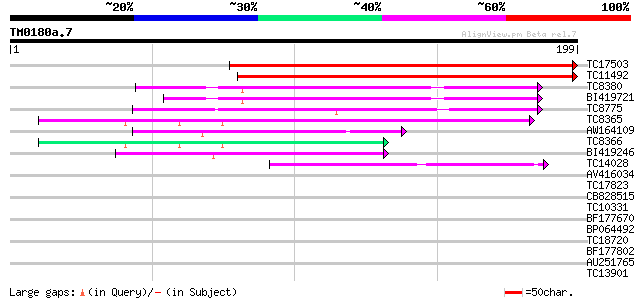
Score E
Sequences producing significant alignments: (bits) Value
TC17503 243 1e-65
TC11492 233 1e-62
TC8380 63 4e-11
BI419721 57 3e-09
TC8775 56 4e-09
TC8365 similar to UP|O24088 (O24088) MtN24 protein, partial (31%) 54 1e-08
AW164109 49 4e-07
TC8366 weakly similar to UP|O24088 (O24088) MtN24 protein, parti... 41 1e-04
BI419246 41 2e-04
TC14028 similar to UP|O24088 (O24088) MtN24 protein, partial (35%) 39 5e-04
AV416034 35 0.007
TC17823 similar to UP|Q93XN3 (Q93XN3) Salicylic acid-induced fra... 35 0.011
CB828515 32 0.056
TC10331 similar to UP|Q8GZT8 (Q8GZT8) PTEN-like protein, partial... 31 0.13
BF177670 31 0.13
BP064492 31 0.16
TC18720 weakly similar to UP|SCWA_YEAST (Q04951) Probable family... 28 0.82
BF177802 28 1.1
AU251765 27 1.8
TC13901 similar to UP|Q9FYU7 (Q9FYU7) UDP-glucose:sinapate gluco... 27 1.8
>TC17503
Length = 564
Score = 243 bits (621), Expect = 1e-65
Identities = 122/122 (100%), Positives = 122/122 (100%)
Frame = +3
Query: 78 DTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASA 137
DTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASA
Sbjct: 3 DTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASA 182
Query: 138 AGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRF 197
AGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRF
Sbjct: 183 AGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRF 362
Query: 198 HL 199
HL
Sbjct: 363 HL 368
>TC11492
Length = 789
Score = 233 bits (595), Expect = 1e-62
Identities = 117/119 (98%), Positives = 118/119 (98%)
Frame = +2
Query: 81 RFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGT 140
RFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGT
Sbjct: 2 RFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGT 181
Query: 141 AMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRFHL 199
AMVRTLKE DTCTASNAFC+QSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRFHL
Sbjct: 182 AMVRTLKEMDTCTASNAFCLQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRFHL 358
>TC8380
Length = 926
Score = 62.8 bits (151), Expect = 4e-11
Identities = 38/146 (26%), Positives = 76/146 (52%), Gaps = 3/146 (2%)
Frame = +3
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTF---RFVLAANAIVAVYSLFEMVAS 101
L R ++ FSL S + + +N HG W +D + R++LA + Y+ ++
Sbjct: 294 LGLRGLALFFSLISLILVASNKHGG----WADFDKYPQYRYLLAIAILSCFYTGGQVHLG 461
Query: 102 VWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ 161
V E+S G + + V FDF DQ+ AYLL++++++ + T++E N F
Sbjct: 462 VHELSTGRKMLQPRIAVLFDFIGDQIMAYLLISSASSAIPITDTIRE----GGDNIFTDS 629
Query: 162 SDIAVALGYAGFVFLGFTSLLTGFRV 187
S A+++ + F+ L +++++G ++
Sbjct: 630 SSSAISMSFLAFICLALSAIISGHKL 707
>BI419721
Length = 527
Score = 56.6 bits (135), Expect = 3e-09
Identities = 34/136 (25%), Positives = 70/136 (51%), Gaps = 3/136 (2%)
Frame = +1
Query: 55 SLASSVFMLTNTHGSDSPHWYHYDTF---RFVLAANAIVAVYSLFEMVASVWEISRGATV 111
SL + + +N HG W +D + R++LA + Y+ ++ V E+S G +
Sbjct: 73 SLDQVLLVASNKHGG----WADFDKYPQYRYLLAIAILSCFYTGGQVHLGVHELSTGRKM 240
Query: 112 FPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYA 171
+ V FDF DQ+ AYLL++++++ + T++E N F S A+++ +
Sbjct: 241 LQPRIAVLFDFIGDQIMAYLLISSASSAIPITDTIRE----GGDNIFTDSSSSAISMSFL 408
Query: 172 GFVFLGFTSLLTGFRV 187
F+ L +++++G ++
Sbjct: 409 AFICLALSAIISGHKL 456
>TC8775
Length = 810
Score = 56.2 bits (134), Expect = 4e-09
Identities = 36/146 (24%), Positives = 75/146 (50%), Gaps = 2/146 (1%)
Frame = +2
Query: 44 ILVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVW 103
+L R+I+F S S + M +N HG + Y+ FR +LA I ++Y+ +++ V
Sbjct: 209 LLWLRVIAFVSSFISFITMASNKHGGWKD-FDKYEEFRHLLATAIISSLYTGYQVFRQVQ 385
Query: 104 EISRGATVFP--EVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ 161
E+ + + + F F DQV AYL++++ ++G M ++ + N F
Sbjct: 386 ELYTEINLLRPRSAVLITFSFVGDQVMAYLMMSSVSSGIPMTSGMRSVE----DNIFTDS 553
Query: 162 SDIAVALGYAGFVFLGFTSLLTGFRV 187
A+++ + F+ L ++L++G+++
Sbjct: 554 LAAAISMSFFAFLCLALSALISGYKL 631
>TC8365 similar to UP|O24088 (O24088) MtN24 protein, partial (31%)
Length = 1003
Score = 54.3 bits (129), Expect = 1e-08
Identities = 46/189 (24%), Positives = 78/189 (40%), Gaps = 15/189 (7%)
Frame = +2
Query: 11 GADGGDNNNNHRSSSPRFHSTMAEHKLRR----FNSLILVFRLISFSFSLAS-------- 58
G +GGD S F + E K R V LISF+ A
Sbjct: 164 GTEGGDGGGGDDGSVSGFLMSKKEEKWCRALLGVRISAFVLCLISFTVMAADKVEGVVIE 343
Query: 59 SVFMLTNTHGSDSPH---WYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEV 115
S+ + N ++ P+ WY Y F++ LA N I VYS ++ V +
Sbjct: 344 SMDLFGNFFSTEEPYSFRWYDYKEFKYSLAVNVIGFVYSGLQICDLVKYLITKKHTLNLR 523
Query: 116 LQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVF 175
++ +F DQ FAYLL++AS++ + + + + F + ++AL + F+
Sbjct: 524 IRGYFSVAMDQGFAYLLMSASSSAASTIHISRSYWVGEGVHKFTDMASASIALSFLAFLA 703
Query: 176 LGFTSLLTG 184
S+++G
Sbjct: 704 FASASIVSG 730
>AW164109
Length = 388
Score = 49.3 bits (116), Expect = 4e-07
Identities = 30/102 (29%), Positives = 54/102 (52%), Gaps = 6/102 (5%)
Frame = +3
Query: 44 ILVFRLISFSFSLASSVFMLTNT------HGSDSPHWYHYDTFRFVLAANAIVAVYSLFE 97
I + R+++F F L + + TN +G + + +R++++ A+ VY+L +
Sbjct: 63 IFLLRVLTFIFLLIALIVTATNKLTQVTINGPREVKFKDFGGYRYMVSTTAVGFVYNLLQ 242
Query: 98 MVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAG 139
M S+ + G F V + FDF D++ +YLLL+ SAAG
Sbjct: 243 MTLSICTLVSGNRPFGNVSYI-FDFFGDKIISYLLLSGSAAG 365
>TC8366 weakly similar to UP|O24088 (O24088) MtN24 protein, partial (28%)
Length = 612
Score = 41.2 bits (95), Expect = 1e-04
Identities = 38/138 (27%), Positives = 55/138 (39%), Gaps = 15/138 (10%)
Frame = +1
Query: 11 GADGGDNNNNHRSSSPRFHSTMAEHKLRR----FNSLILVFRLISFSFSLAS-------- 58
G +GGD S F + E K R V LISF+ A
Sbjct: 199 GTEGGDGGGGDDGSVSGFLMSKKEEKWCRALLGVRISAFVLCLISFTVMAADKVEGVVIE 378
Query: 59 SVFMLTNTHGSDSPH---WYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEV 115
S+ + N ++ P+ WY Y F++ LA N I VYS ++ V +
Sbjct: 379 SMDLFGNFFSTEEPYSFRWYDYKEFKYSLAVNVIGFVYSGLQICDLVKYLITKKHTLNLR 558
Query: 116 LQVWFDFGHDQVFAYLLL 133
++ +F DQ FAYLL+
Sbjct: 559 IRGYFSVAMDQGFAYLLM 612
>BI419246
Length = 563
Score = 40.8 bits (94), Expect = 2e-04
Identities = 25/106 (23%), Positives = 46/106 (42%), Gaps = 10/106 (9%)
Frame = +1
Query: 38 RRFNSLILVFRLISFSFSLASSVFMLTNTHGSD----------SPHWYHYDTFRFVLAAN 87
RR ++ L R + + S+ S M+T S W +++ +++ +
Sbjct: 244 RRAEAIQLSLRALCMATSVVSLSLMVTAKQASSVSIYGFRLPVHSKWSFSESYEYLVGVS 423
Query: 88 AIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLL 133
A VA +S +++ R +++ P W F DQVFAY L+
Sbjct: 424 AAVAAHSFLQLLIGSSRFLRMSSLIPSRNHAWLIFAGDQVFAYALM 561
>TC14028 similar to UP|O24088 (O24088) MtN24 protein, partial (35%)
Length = 532
Score = 39.3 bits (90), Expect = 5e-04
Identities = 26/98 (26%), Positives = 47/98 (47%)
Frame = +2
Query: 92 VYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDT 151
VYS ++ +S G V L+V+F F DQ+ YLL++AS++ + +
Sbjct: 11 VYSGLQICDLGLYLSTGKHVVEHRLRVYFSFALDQILTYLLMSASSSAAPRA---YDWQS 181
Query: 152 CTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVC 189
+ F ++ +VAL + F SL++G ++C
Sbjct: 182 HWGEDQFPYMANASVALSFVAFAAFALASLVSG-SIIC 292
>AV416034
Length = 392
Score = 35.4 bits (80), Expect = 0.007
Identities = 37/133 (27%), Positives = 57/133 (42%), Gaps = 4/133 (3%)
Frame = +2
Query: 3 RSPQPLRNG--ADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSV 60
RSP L G A GG+ + +++ R SL RL+ +++ V
Sbjct: 50 RSPMQLMMGTAASGGEELEGNGATTLR--------------SLETFLRLLPIGLCVSALV 187
Query: 61 FMLTNTHGSD--SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQV 118
ML N+ +D S + FR+++ AN I A YSL V + R + P + +
Sbjct: 188 LMLKNSQENDYGSVDYTDLGPFRYLVHANGICAGYSLLSAVFVA--VPRPS---PTMSRA 352
Query: 119 WFDFGHDQVFAYL 131
W F DQV Y+
Sbjct: 353 WTFFLLDQVLTYI 391
>TC17823 similar to UP|Q93XN3 (Q93XN3) Salicylic acid-induced fragment 1
protein (Fragment), partial (32%)
Length = 363
Score = 34.7 bits (78), Expect = 0.011
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 2/89 (2%)
Frame = +3
Query: 48 RLISFSFSLASSVFMLTNTHGSDSPHWYHYD--TFRFVLAANAIVAVYSLFEMVASVWEI 105
RL + + V ML N+ ++ + D FR+++ AN I A YSLF V + +
Sbjct: 114 RLFPIGLCVTALVIMLKNSQENEYGTLDYTDLSAFRYLVHANGICAGYSLFSAV--IIAL 287
Query: 106 SRGATVFPEVLQVWFDFGHDQVFAYLLLA 134
R +T + + W F DQ+ Y++LA
Sbjct: 288 PRPST----MPRAWTFFVLDQLLTYIILA 362
>CB828515
Length = 496
Score = 32.3 bits (72), Expect = 0.056
Identities = 17/64 (26%), Positives = 34/64 (52%)
Frame = -3
Query: 126 QVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 185
Q AYLL++AS++ + + + T ++ F + AVAL + F+ S+++G
Sbjct: 314 QGLAYLLMSASSSAASTIHIYRSYWTGEGAHTFTDMASAAVALSFLAFLAFASASIVSGL 135
Query: 186 RVVC 189
++C
Sbjct: 134 -ILC 126
>TC10331 similar to UP|Q8GZT8 (Q8GZT8) PTEN-like protein, partial (7%)
Length = 605
Score = 31.2 bits (69), Expect = 0.13
Identities = 22/71 (30%), Positives = 30/71 (41%), Gaps = 5/71 (7%)
Frame = -2
Query: 117 QVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTAS-----NAFCVQSDIAVALGYA 171
+ W +FG D + + A A V L E C+AS ++F SD +A G A
Sbjct: 217 ETWSEFGFDSHSLGAFVTVATASDAWVCLLLEEAGCSASPSENTSSFSAPSDSLLAFGAA 38
Query: 172 GFVFLGFTSLL 182
G G LL
Sbjct: 37 GLTSTGAGLLL 5
>BF177670
Length = 341
Score = 31.2 bits (69), Expect = 0.13
Identities = 26/91 (28%), Positives = 41/91 (44%), Gaps = 7/91 (7%)
Frame = +1
Query: 102 VWEISRGATVFPEVLQVWFDFGHDQVFAYLLLA--ASAAGTAMVRTLKETD-----TCTA 154
V + RG +F + L W F DQV AY+ +A A+AA +AM L + + C
Sbjct: 19 VLSMIRGKVLFSKPL-AWAIFSGDQVMAYITVAAVAAAAQSAMFAKLGQPELQWMKICDM 195
Query: 155 SNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 185
FC Q +A + + + S ++ F
Sbjct: 196 YGKFCNQVGEGLASAFLASLSMVVLSCISAF 288
>BP064492
Length = 482
Score = 30.8 bits (68), Expect = 0.16
Identities = 14/43 (32%), Positives = 26/43 (59%)
Frame = -1
Query: 106 SRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKE 148
S G + + V FDF DQ+ AYLL++++++ + T++E
Sbjct: 482 SPGRKMLQPRIAVLFDFIGDQIMAYLLISSASSAIPITDTIRE 354
>TC18720 weakly similar to UP|SCWA_YEAST (Q04951) Probable family 17
glucosidase SCW10 precursor (Soluble cell wall protein
10) , partial (7%)
Length = 871
Score = 28.5 bits (62), Expect = 0.82
Identities = 22/62 (35%), Positives = 34/62 (54%)
Frame = +3
Query: 132 LLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFI 191
++ +SAA +VRTL+ +D T+S A + D AL Y +F G S + +V F+
Sbjct: 429 IIGSSAAVPFLVRTLQSSDVKTSSQA---KQDALRAL-YNLSIFPGNISFILETHLVPFL 596
Query: 192 IN 193
IN
Sbjct: 597 IN 602
>BF177802
Length = 481
Score = 28.1 bits (61), Expect = 1.1
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = -2
Query: 54 FSLASSVFMLTNTHGSDSPHW 74
FSL SVF+ +NTH +DSP W
Sbjct: 105 FSL--SVFL*SNTHSTDSPSW 49
>AU251765
Length = 332
Score = 27.3 bits (59), Expect = 1.8
Identities = 16/41 (39%), Positives = 24/41 (58%)
Frame = +2
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLA 85
L R I+ FSL S + M +N HG D + Y+ +R++LA
Sbjct: 209 LGLRGIALLFSLISFIAMASNQHG-DWREFDKYEEYRYLLA 328
>TC13901 similar to UP|Q9FYU7 (Q9FYU7) UDP-glucose:sinapate
glucosyltransferase , partial (16%)
Length = 558
Score = 27.3 bits (59), Expect = 1.8
Identities = 14/42 (33%), Positives = 20/42 (47%)
Frame = +2
Query: 7 PLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFR 48
P G GG ++N+HR S +T RRF + L+ R
Sbjct: 155 PKGRGDGGGSSSNHHRHSQRNIITTTTHTNRRRFPPVRLIRR 280
Score = 25.0 bits (53), Expect = 9.0
Identities = 12/33 (36%), Positives = 16/33 (48%)
Frame = +2
Query: 7 PLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRR 39
P + DGG +++NH S R T H RR
Sbjct: 152 PPKGRGDGGGSSSNHHRHSQRNIITTTTHTNRR 250
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.328 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,789,099
Number of Sequences: 28460
Number of extensions: 50273
Number of successful extensions: 402
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 399
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 401
length of query: 199
length of database: 4,897,600
effective HSP length: 86
effective length of query: 113
effective length of database: 2,450,040
effective search space: 276854520
effective search space used: 276854520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0180a.7


